INTRODUCTION
Sera and other body fluids contain cell free DNA,RNA and circulating nucleic acids which serve as potential biomarkers [1]. One such biomarker which has aroused interest in recent years is a small non-coding (18 – 24 ~nt) RNA which is called as microRNA (miRNA) [2]. It was discovered in 1993 by Ambros et al., in Caenorhabditis elegans [3]. The important roles of miRNA are regulation of the gene expression, cell-cell communication, cell division and apoptosis [2]. Recent data show that there are over 1500 matured and primary miRNA(pri miRNA) which are found in humans, with certain definite functions.
The MiRNAs are transcribed similarly as protein coding genes by RNA polymerase II [4] and RNA polymerase III [5]. The pri-miRNA is transcribed in the nucleus and undergoes post transcriptional processing in the cytoplasm, which is similar to that of other RNAs. DROSHA and DICER are RNase III enzymes which are involved in both the phases. During its processing, the ~70nt primiRNA is converted into 18 – 24 ~nts matured double strand miRNA (ds miRNA) [Table/Fig-1 & 2] [6].
Biogenesis of Pathway of miRNA
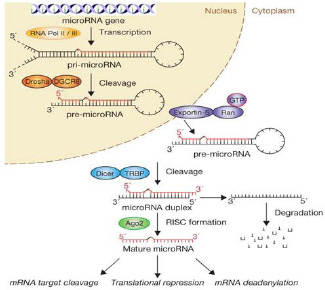
One of the functions of miRNA is downregulation of the gene expression, which makes it a useful diagnostic and prognostic tool in cancer [7]. It has high stability in blood, which enables an easy storage of the sample for the estimation of miRNA [8]. The current studies have provided a new algorithm for the screening and diagnosis of various diseases by using miRNA. This review pertains to the whole process of biogenesis, secretion, regulation and the use of miRNA in health and diseases.
THE CURRENT STATUS OF MIRNA AS A BIOMARKER
Biomarkers may be defined as biological substances which are more specific and sensitive to a particular physiological or a pathological condition [Table/Fig-3]. The present studies show that miRNA fits into all the criteria for biomarkers [Table/Fig-4]. It is stable in various body fluids; the sequences of most of the miRNAs are conserved among different species; the expressions of some miRNAs are tissue specific and the biological conditions as well as the levels of miRNAs can be assessed by using modern methods. Recently, we use specific proteins, enzymes and isoenzymes as biomarkers for the diagnosis of many a disease.
MiRNAs in cell differentiation and development
| Let -7 – miRNAs family | Stabilizes the self-renewing versus differentiated cell fate |
| Bantam miRNA | Physically associated with dFmrp in the ovary. It acts as an extrinsic factor for germline stem cell maintenance. Bantam interacts genetically with dFmr1 to regulate the fate of germ line stem cells |
| miRNA – 223 | Plays an important role in differentiation and function of osteoclasts. Decreased osteoclast number and bone resorption. It regulates NF1 – A and H – CSFn levels |
| miRNA- 128 | Overexpression in glioma cells inhibits cell proliferation. Target site is within the 3’UTR of E2F3a, a transcription factor which regulates cell cycle progression. The protein level of E2F3a negatively correlates with miR-128 |
Characteristics of an ideal biomarker
| Specific | Specific tissue. |
| Sensitive | Can be differentiated in all pathological conditions. Early and significant variation in levels during the different stages of disorders. |
| Predictive | Long half-life. |
| Robust | Easily identified and quantified using modern techniques. |
| Translatable | Data can be used to bridge pre-clinical and clinical gap |
| Non invasive | Present in circulation. |
The examples include troponin in the diagnosis of myocardial infarction, Prostate Specific Antigen (PSA) for prostate cancer and Alanine Aminotransferase (ALT) and Aspartate Aminotransferase (AST) enzymes for the liver function, etc [6].
The serum miRNA studies began with the associated levels of miR-21 in patients with diffuse large B-cell lymphoma. The expression of the miRNA profiles for lung cancer [Table/Fig-5], colorectal cancer, Diabetes mellitus and other diseases has also assumed relevance. Any biological sample can be used to assay the miRNA. Presently, the circulating miRNAs are easy to detect and to quantitate.
MiRNA expression and tumor genesis
| miR-15 & miR-16 | Absent or downregulation in chronic lymphocytic leukemia |
| miR-34a | Absence of miR-34a is seen in pancreatic cancer cell, induced by p53 tumor suppressor protein. Involved in cell cycle progression, apoptosis, DNA repair and angiogenesis. Its expression is silenced and attributed to aberrant CPG methylation of its promoter. |
| miR-23b | Repressed in Acute Myelogenous leukemia (AML) |
| miR-155 | Increased expression in AML |
In prostate cancer, the level of miR-142 is significant, as compared to that in healthy individuals [9]. In urine, the miR-126 and the miR-182 ratio helps in the diagnosis of bladder cancer [10]. In saliva, low levels of miR-125a and -200a detect oral squamous cell carcinoma [11].
MIRNAS AND CANCER
Previously, carcinogenesis was attributed to abnormalities in the oncogenes and the tumour-suppressing genes. Recently, it was recognized that miRNA also possesses an important role in the diagnosis and prognosis of cancer. The links between the dysregulation and the reduced expression of the tumour suppressor genes in cancer were shown in earlier studies [12]. Calin et al., located miR-15 and miR-16 on chromosome 13q14, a region that is deleted in more than half of the patients of chronic lymphocytic leukaemia and B-cell leukaemia [13]. The overexpression of the let 7 family on the RAS oncogenes lead to lung cancer [14]. MiRNAs are also called ‘oncomirs’, because both in in vitro and in vivo conditions, they can be viewed during the cell growth, cell cycle progression and during the invasion in all types of cancers [15]. A series of studies (He et al., Lu et al.,; O’Donnell et al.,) has reported the association of miRNAs and cancer [16–18]. The dysregulation of a group of miRNAs resulted in overexpression of the MYC oncogenes, which is associated with B-cell neoplasia [12].
A study which was performed by Arndt et al., showed a significantly low level of expression of miR-143 and miR-145 in colorectal carcinoma [19]. Similarly, miR-10b, miR-125b and -145 are downregulated and miR-21 and miR-155 are upregulated in breast cancer [20]. Among the miRNAs, miR-145 is involved in downregulation. Moreover, it helps in finding out the progression of the normal breast tissues into cancerous tissues. The Mi-21 expression is progressively upregulated during the progression of breast cancer. Some of them are associated with the invasion and the prognosis of breast cancer [21]. Tumours are classified, based on their miRNA expression, which can be used in their diagnosis and prognosis [Table/Fig-6].
miRNAas putative biomarker in different pathological conditions
| Condition | MiRNAs |
| Diffuse large B-cell lymphoma | Increased levels of miR-155, miR-210 and miR-21 |
| Prostate cancer | Distinguishing level of miR-144 |
| Breast cancer | Increased levels of miR-10b and 34a Decreased levels of miR-195, let-7a |
| Ovarian cancer | Increased levels of miR-21, -141, -200a, 200b, 200c, 203 205 and miR-214 |
| Colorectal cancer | Increased levels of miR-29a, -17-3p and miR-92a (Plasma) |
| Bladder cancer | Ratio of urinary miR-126 and miR-182 |
| Oral cancer | Increased levels of miR-31 (Plasma) and miR-125a, -200a (saliva) |
| Myocardial Infarction | Increased levels of miR-499 and miR-208b |
| Congestive Heart Failure | Increased levels of miR-423-5p |
| Coronary Artery Disease | Decreased levels of miR-126, cluster miR-17/92 and miR-155a |
MIRNAS IN THE NERVOUS SYSTEM (NS) REGULATION
MiRNAs play an important role in maintaining the survival of the mature neurons and their functions. MiRNA-134 contributes to the synaptic development, maturation and plasticity. A defective expression of miR-134 could be associated with diseases viz., Alzheimer’s disease, the Fragile-X-Syndrome (FXS) and autism [22]. MiR-133b regulates the maturation and the function of the midbrain dopaminergic neurons and it is deficient in patients with Parkinson’s disease [23]. MiR-8 directly targets atrophin and the miR-8 mutant phenotypes are attributable to the enhancement of the datrophin activity, resulting in elevated apoptosis in the brain and behavioural defects. MiR-124 promotes the NS development by regulating an intricate network of NS specific alternative splicing. Mi-124 also affects certain regulators of the neuro-specific gene expression [24].
MiRNA-124 directly inhibits the small C-terminal domain phosphatase- 1 (SCP-1) which is required for the RE1 silencing transcription factor (REST) mediated repression of the neuronal genes [25]. When cells differentiate to neurons, the REST transcription is inhibited and miR-124 promotes the elimination of the biological effects of REST by inhibiting SCP1 [26].
MIRNAS AND VIRAL INFECTIONS
The host cells use miRNAs to target certain essential viral functions. In turn, the viruses use miRNAs to control their host cells. An antiviral miRNA against the retovirus primate foamy virus type I (PFV – I) in human cells was the first to be reported [27]. The SV 40 encoded miRNA -miR-S1 helps in keeping the infected cell hidden from the immune system. It is expressed late in the viral replication cycle and it helps in degrading the viral mRNA encoding T antigen. This limits the exposure of the infected cell to the cytotoxic ‘T’ lymphocytes [28]. The expression of the host cell miR-122 can inhibit the replication of the hepatitis C virus and it works through IFN – ß. The miRNA silencing machinery plays a physiological role in controlling the HIV – 1 replication [29] .
MIRNAS AND THE CARDIOVASCULAR SYSTEM
Coronary heart disease is still the leading cause of death worldwide. Recent studies have indicated that several miRNAs are dysregulated in CAD patients. In cardiovascular diseases, several specific circulating miRNAs have been profiled as novel biomarkers for distinguishing among the different cardio vascular events. Studies which had been performed in early 2008 have demonstrated the link between the circulating miRNAs and diseases other than cancer [8,9,30].
Adachi et al reported that miR-499 was elevated in acute myocardial infraction, but that it was not elevated in coronary heart disease [31]. Wang et al presented miR-208a as a sensitive, early marker of AMI, because the levels got elevated within 4 hrs [32]. Corsten et al., observed significant correlations concerning the levels of miR- 208b and miR-499 with the serum levels of troponin T and Creatine Kinase (CK), which are two established markers of cardiac injuries [33].
Tijisen et al., studied 108 miRNAs by using microarray and reported that they were differentially expressed in coronary heart failure patients. A further analysis which was performed by RT-qPCR showed a rise in miR-423-5p in other Congestive Heart Failure (CHF) patients. It had a correlation with the N-terminal pro-hormone brain natriuretic peptide levels (NT-proBNP) and the ejection fraction, suggesting that it would be a good predictor of the CHF diagnosis [34].
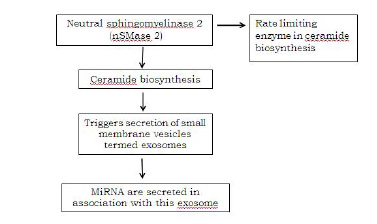
Fichtlscherer et al., in the plasma of CAD patients, detected low levels of the miRNAs, namely miR-126, the miR17/92 cluster and miR-208b and also found them in low levels at the baseline. A prospective evaluation of the data showed low levels of miR -126, -17, 92a and miR-155. The MiRNAs which were packed in exosomes, microvesicles or apoptotic bodies, which were taken up by atherosclerotic lesions, led to a speculated reduction in the levels of the circulating miRNAs in CAD patients [35].
MIRNAS AND GASTROINTESTINAL DISEASES
Recently, it was identified that 22 miRNAs were regulated and that 13 were downregulated in gastric cancers [36]. MiR-15b and miR-16 were down regulated in human gastric cancer cells [37]. They act by modulating apoptosis. A considerable upregulation of miR-135a and miR-135b was observed in colorectal adenomas and carcinomas and it was correlated significantly with the low Adenomatous Polyposis Coli(APC) mRNA levels. MiR-135a and miR- 136b target the 3’UTR of the APC gene and cause its suppression. Thus, microRNAs are very much associated with the pathogenesis of colorectal cancers [38].
MIRNAS AND DIABETES MELLITUS
MiR-7 and miR-375 are expressed in high levels in the pancreas. They are associated with both the development of the pancreas and also the secretion of insulin. MiR-375 directly targets 3’ phosphoinositide- Dependent Protein Kinase – 1 (PDK-1) and it decreases the glucose stimulatory action on the insulin gene expression [39]. Glucose decreases the miR-375 precursor level and a concomitant increase in the PDK-I protein [40]. The biological mechanism of a recently discovered association of type 2 diabetes with an ACAA insertion/deletion polymorphism at 3’ UTR of the IGF2R gene can be very well explained on the basis of the role of miRNA [41]. Decreased levels of miR-20b, miR-21, miR-24, miR- 15a, miR-126, miR-191, miR-197, miR-223, miR-320 and miR- 486 were significant in diabetes, but there could be a moderate increase in miR-28-3p [35].
THE ANALYSIS OF MIRNA
Recent studies have depicted that miRNAs could be used as noninvasive markers for the diagnosis and prognosis of conditions like acute myocardial infarction, congestive heart failure, cancer and drug induced liver damage [32,34,42]. miRNAs showed resistance towards their degradation by RNase, which made them a promising diagnostic tool as compared to the miRNAs [8,9,43,44].
Only, few data are available which pertain to the analytical assays of miRNAs. Jennifer et al discussed the pre-analytical and the analytic errors of the circulating miRNAs. The concentrations of the miRNAs will be altered by pre-analytical factors like the protocol of the sample collection, centrifugation (rpm) and stability. There will be an increase in the total miRNA concentration due to contamination of the sample with platelets, erythrocytes and haemolysis. Usually, miRNA is stable for 24hrs at room temperature and for 72hrs when it stored at – 4°C [45].
The quantification of miRNA is performed both in plasma and serum. Both the serum and the plasma concentrations of the miRNAs are different. As compared to plasma, serum depicts a good correlation as a marker, even though serum contains a low concentration of miRNA. Commercial kits are available for the extraction of miRNA from other body fluids [46].
A spectrophotometer rarely determines small amounts of miRNA. Alternative modern techniques such as Real Time PCR (RT-PCR) for qualitative identification and qPCR can estimate its concentration by measuring the electrical signals which are formed from the oxidation of guanine which is induced by the hybrid formation of miRNA [47]. The microarray technique provides functional genomics through the parallel expression measurements of the genomes, which could be used in the research which pertains to drug discovery and the targeting and the determination of the biomarkers. Microarray enables the quantitation and the sequencing of miRNA and this could ultimately be acceded into the database [48].
[1]. Swarup V, Rajeswari MR, Circulating (cell-free) nucleic acids--a promising, non-invasive tool for early detection of several human diseasesFEBS Lett. 2007 Mar 6 581(5):795-99. [Google Scholar]
[2]. Bartel DP, MicroRNAs: genomics, biogenesis, mechanism, and functionCell 2004 Jan 23 116(2):281-97. [Google Scholar]
[3]. Lee RC, Feinbaum RL, Ambros V, The C. elegansheterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14Cell 1993 Dec 3 75(5):843-54. [Google Scholar]
[4]. Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, MicroRNA genes are transcribed by RNA polymerase IIThe EMBO Journal 2004 Sep 16 23(20):4051-60. [Google Scholar]
[5]. Borchert GM, Lanier W, Davidson BL, RNA polymerase III transcribes human microRNAsNat. Struct. Mol. Biol. 2006 Dec 13(12):1097-101. [Google Scholar]
[6]. Etheridge A, Lee I, Hood L, Galas D, Wang K, Extracellular microRNA: A new source of biomarkersMutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 2011 Dec 717(1-2):85-90. [Google Scholar]
[7]. Kosaka N, Iguchi H, Ochiya T, Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosisCancer Science 2010 Jul 7 101(10):2087-92. [Google Scholar]
[8]. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseasesCell Research 2008 Oct 18(10):997-1006. [Google Scholar]
[9]. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Circulating microRNAs as stable blood-based markers for cancer detectionProceedings of the National Academy of Sciences 2008 Jul 28 105(30):10513-18. [Google Scholar]
[10]. Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancerUrol. Oncol. 2010 Dec 28(6):655-61. [Google Scholar]
[11]. Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer DetectionClinical Cancer Research 2009 Aug 25 15(17):5473-77. [Google Scholar]
[12]. Palmero EI, Campos SGP de, Campos M, Souza NCN de, Guerreiro IDC, Carvalho AL, Mechanisms and role of microRNA deregulation in cancer onset and progressionGenetics and Molecular Biology 2011 34(3):363-70. [Google Scholar]
[13]. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemiaProc. Natl. Acad. Sci. U.S.A. 2002 Nov 26 99(24):15524-29. [Google Scholar]
[14]. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, RAS is regulated by the let-7 microRNA familyCell 2005 Mar 11 120(5):635-47. [Google Scholar]
[15]. Wurz K, Garcia RL, Goff BA, Mitchell PS, Lee JH, Tewari M, MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: Relationship to CDKN1B, CDKNIC and overall survivalGenes Chromosomes Cancer 2010 Jul 49(7):577-84. [Google Scholar]
[16]. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, A microRNA polycistron as a potential human oncogeneNature 2005 Jun 9 435(7043):828-33. [Google Scholar]
[17]. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, MicroRNA expression profiles classify human cancersNature 2005 Jun 9 435(7043):834-38. [Google Scholar]
[18]. O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT, c-Mycregulated microRNAs modulate E2F1 expressionNature 2005 Jun 9 435(7043):839-43. [Google Scholar]
[19]. Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancerBMC Cancer 2009 9:374 [Google Scholar]
[20]. Blenkiron C, Miska EA, miRNAs in cancer: approaches, aetiology, diagnostics and therapyHuman Molecular Genetics 2007 Apr 15 16(R1):R106-R113. [Google Scholar]
[21]. Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X, Evaluation of Genetic Variants in MicroRNA-Related Genes and Risk of Bladder CancerCancer Research 2008 Apr 1 68(7):2530-37. [Google Scholar]
[22]. Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V, Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiationGenome Biol. 2004 5(3):R13 [Google Scholar]
[23]. Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG, Regulation of miRNA expression during neural cell specificationEur. J. Neurosci. 2005 Mar 21(6):1469-77. [Google Scholar]
[24]. Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ, Biological functions of microRNAs: a reviewJournal of Physiology and Biochemistry 2010 Oct 28 67(1):129-39. [Google Scholar]
[25]. Coulson JM, Transcriptional regulation: cancer, neurons and the RESTCurr. Biol. 2005 Sep 6 15(17):R665-68. [Google Scholar]
[26]. Shi Y, Jin Y, MicroRNA in cell differentiation and developmentSci. China, C, Life Sci. 2009 Mar 52(3):205-11. [Google Scholar]
[27]. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, A cellular microRNA mediates antiviral defense in human cellsScience 2005 Apr 22 308(5721):557-60. [Google Scholar]
[28]. Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D, SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cellsNature 2005 Jun 2 435(7042):682-86. [Google Scholar]
[29]. Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Suppression of microRNA-silencing pathway by HIV-1 during virus replicationScience 2007 Mar 16 315(5818):1579-82. [Google Scholar]
[30]. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphomaBr. J. Haematol. 2008 May 141(5):672-75. [Google Scholar]
[31]. Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, Plasma microRNA 499 as a biomarker of acute myocardial infarctionClin. Chem. 2010 Jul 56(7):1183-85. [Google Scholar]
[32]. Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humansEuropean Heart Journal 2010 Feb 16 31(6):659-66. [Google Scholar]
[33]. Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular diseaseCircCardiovasc Genet 2010 Dec 3(6):499-506. [Google Scholar]
[34]. Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, MiR423-5p as a circulating biomarker for heart failureCirc. Res. 2010 Apr 2 106(6):1035-39. [Google Scholar]
[35]. Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Circulating microRNAs in patients with coronary artery diseaseCirc. Res. 2010 Sep 3 107(5):677-84. [Google Scholar]
[36]. Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysisLancet Oncol. 2010 Feb 11(2):136-46. [Google Scholar]
[37]. Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cellsInt. J. Cancer. 2008 Jul 15 123(2):372-79. [Google Scholar]
[38]. Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JAF, Bolijn A, Regulation of the Adenomatous Polyposis Coli Gene by the miR-135 Family in Colorectal CancerCancer Research 2008 Jul 15 68(14):5795-802. [Google Scholar]
[39]. El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E, miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cellsDiabetes 2008 Oct 57(10):2708-17. [Google Scholar]
[40]. Joglekar MV, Parekh VS, Hardikar AA, Islet-specific microRNAs in pancreas development, regeneration and diabetesIndian J. Exp. Biol. 2011 Jun 49(6):401-08. [Google Scholar]
[41]. Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S, Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetesBiochem.Biophys. Res. Commun. 2008 Sep 12 374(1):101-05. [Google Scholar]
[42]. Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Circulating microRNAs, potential biomarkers for drug-induced liver injuryProceedings of the National Academy of Sciences 2009 Feb 25 106(11):4402-07. [Google Scholar]
[43]. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Serum MicroRNAs Are Promising Novel Biomarkers. Williams S, editorPLoS ONE 2008 Sep 5 3(9):e3148 [Google Scholar]
[44]. Chim SSC, Shing TKF, Hung ECW, Leung Ty., Lau Tk, Chiu RWK, Detection and Characterization of Placental MicroRNAs in Maternal PlasmaClinical Chemistry 2008 Mar 1 54(3):482-90. [Google Scholar]
[45]. McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A, Analysis of Circulating MicroRNA: Preanalytical and Analytical ChallengesClinical Chemistry 2011 Apr 12 57(6):833-40. [Google Scholar]
[46]. Jan CB, Daniela Wuttig, Ruprecht Kuner, Holger Sultmann, serum microRNAs as non-invasive biomarkers for cancerMolecular cancer [Google Scholar]
[47]. Li J, Yao B, Huang H, Wang Z, Sun C, Fan Y, Real-time polymerase chain reaction microRNA detection based on enzymatic stem-loop probes ligationAnal. Chem. 2009 Jul 1 81(13):5446-51. [Google Scholar]
[48]. Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B, Detection of cancer with serum miRNAs on an oligonucleotide microarrayPLoS ONE 2009 4(7):e6229 [Google Scholar]