The UTIs are a prevalent bacterial infection across all ages, particularly common in females due to anatomical factors such as a shorter urethra, its proximity to the anal canal, and the impact of sexual activity [1]. While Escherichia coli (E. coli) is the most frequent culprit and other gram-negative bacteria including Pseudomonas, are implicated in UTIs [2]. The pathogenicity of these uropathogens in UTIs is contingent upon the presence of various virulence factors, including adhesins, capsules, siderophores, toxins. IIB invade urothelium and form IBCs within the bladder epithelium mimicking biofilm-like structures [2]. Females are at a heightened risk of recurrent UTIs within six months to a year following appropriate treatment of the initial infection and subsequent negative urine cultures. Over half of all recurrent episodes of acute uncomplicated cystitis are attributed to the same bacterial strain responsible for the initial infection [3]. These recurrences significantly inflate healthcare costs and diminish quality of life. Recurrences may be due to reinoculation of the bladder lumen by an uropathogenic strain persisting in the periurethral or faecal flora following a previous UTI [4]. Uropathogenic bacteria can establish an intracellular niche within the bladder, facilitated by Type 1 pili that enable binding and invasion of bladder urothelial cells. This pathway aids in replication and the formation of IBCs with biofilm-like properties [4].
The IBC are transient and often adopt a filamentous morphology, enabling evasion of neutrophil engulfment and facilitating re-invasion in the urothelium. During infection, exfoliated urothelial cells are expelled into the urine, allowing bacteria to persist for weeks, shielded from antibiotics and the host immune system. Epithelial turnover may trigger quiescent bacteria to revert to an actively replicative form, resulting in recurrent bacteriuria [5]. However, recurrent or culture negative, undiagnosed UTI with IIB/IBC may lead to chronic cystitis [5]. Identifying intracellular bacteria alongside assessing their virulence genes is crucial for comprehending the pathophysiology of UTIs. These bacteria, resistant to antibiotics, contribute to recurring infections, posing challenges in treatment.
Antibiotic resistance is a global challenge, so to identify recurrent UTI with the uropathogenic strain is essential for precise treatment of UTI. Intracellular Gram-negative bacteria like E. coli and Pseudomonas forms biofilm like structures and become resistant for treatment. Recently, few studies demonstrated the use of confocal microscopy for detection of IIB/IBC in urine samples suspected with UTI, Rosen DA et al., detected almost 50% of IBC/IIB by confocal microscopy which was missed by light microscopy [3]. Confocal microscopy is a high-resolution fluorescence imaging and sensitive technique in which laser light is focused to a diffraction limited spot at a specific depth within the sample [6].
Materials and Methods
This was a cross-sectional observational study done for detection of IIB/IBC in urine samples suspected with UTI. The urine samples were obtained from women who visited for diagnosis of UTI at Central Laboratory of Dr. D. Y. Patil Medical College, Hospital and Research Centre, Pune, Maharashtra, India between January 2022 to December 2022.
The study was approved by the Institutional Ethics Committee (Ref. No. DYPV/EC/46/18) and informed consent was obtained from all the participants.
Inclusion criteria: Women aged 18-50 years who visited for diagnosis of UTI and not receiving antibiotic treatment were included in the study.
Exclusion criteria: Women below 18 and above 50 years, women who have taken antibiotic, women having chronic renal complications and not willing to participate were excluded from the study.
Study Procedure
Four hundred ninety-five urine samples were collected from women of age group 18-50 years. Their demographic information, clinical symptoms, and history of recurrence of UTI were recorded. Samples were collected for a fixed duration of one year. The single time point random urine sample was collected prior to antibiotic treatment as per the standard collection protocol and immediately processed for routine microscopy using Gram and Giemsa staining followed by conventional culture using Cystine-Lactose-Electrolyte-Deficient agar (CLED) medium and biochemical tests like Indole, Methyl Red, Voges Proskauer, Citrate and motility test were done for lactose fermenting pink colonies of E. coli for confirmation. As shown in [Table/Fig-1], 149 urine samples detected with gram negative rods, out of that 92 were processed for immunofluorescence staining by confocal microscopy [7]. From these 92 samples, 24 culture positive urine samples were further examined for virulence factors using real-time PCR, and their biofilm-forming ability was assessed using the tube method [8]. One hundred seven urine samples detected with yeasts and 16 samples with polymicrobial flora were excluded from confocal analysis.
Urine sample selection criteria for detection of intracellular bacteria by Confocal Microscopy.
UTI: Urinary tract infection; CLSM: Confocal laser scanning microscopy; PCR: Polymerase chain reaction
Detection of Intracellular Bacteria
Four hundred and ninety-five samples processed for light microscopy. Random urine sample of volume 1 to 5 mL were centrifuged at a speed of 2000 rpm for a duration of 10 minutes using Eppendorf 5810 R centrifuge [9,10]. The sediment obtained postcentrifugation was employed for Gram and Giemsa staining, which aids in the identification of gram-negative bacteria residing within urothelial cells, as observed under light microscopy [11]. The remaining sediment was promptly fixed using a 4% formaldehyde solution. Following a rinse with Phosphate Buffered Saline (PBS), the sediment was placed on a Poly-l-lysine coated glass slide for fluorescent staining. Concurrently, the same sample was processed for routine culture using CLED medium [12].
Under light microscopy, the heat-fixed slides with sediment were stained using the Gram and Giemsa methods and examined under a light microscope (Lawrence Mayo). The presence of Gram-negative rods and filamentous forms was observed with Gram staining, while Giemsa staining was used to observe darkly stained urothelial cells for potential intracellular communities. Samples demonstrating the presence of intracellular bacteria were further processed for fluorescence staining [13].
In the confocal microscopy, the formalin-fixed cell deposit was placed on a Poly-l-lysine coated glass slide (Rohem double frosted slides). After a PBS wash, the slides were stained with Wheat Germ Agglutinin (WGA) Alexa fluor™ 350 (5.0 ug/mL) and Uroplakin III (1/50) for 15 minutes in a dark room at ambient temperature. Cell permeabilisation was then performed using 0.3% Triton-X 100 for 15 minutes. The slides were subsequently washed with PBS and incubated with an E. coli antibody coupled to FITC (Abcam) (1/50) and Uroplakin III (Santa Cruz Biotechnology) primary antibody for one hour at room temperature. The slides were then washed with PBS for 15 minutes and incubated with a goat antimouse secondary Uroplakin III antibody (1/200) and WGA (1/100) for 30 minutes. After staining, the slides were mounted with 10 μL of ultra-Cruz mounting medium [13].
Pseudomonas were also identified using Gram staining and confirmed by conventional culture using CLED medium and biochemical reactions like Voges Proskauer, indole, and methyl red tests, oxidase and catalase test. Non lactose fermenting colorless colonies of Pseudomonas showing negative Voges Proskauer, indole, and methyl red, but oxidase, catalase positive, and motility test positive were further processed for fluorescence staining. For confocal microscopy, a similar staining protocol was employed for the detection of Pseudomonas in urothelial cells using an anti-Pseudomonas antibody (1/50) from Thermo Fisher Scientific. Acquisition was performed with a confocal laser scanning microscope 710 (Zeiss) using 405 nm diode laser (350/460), 488 nm Argon laser (488/520), and 561 nm diode laser lines (543/797). Z stacks were acquired using a step size of 0.3 um and a pixel size of 1024×1024 in the x-y plane. Fiji software was utilised for the analysis of the images [13].
Biofilm Detection by Tube Method
For the detection of biofilm, the tube method was employed. A urine sample demonstrating the presence of intracellular bacteria was inoculated in 1 mL of Trypticase soya broth in polystyrene tubes and incubated for 24 hours at 37° Celsius. After overnight incubation, the turbid broth was discarded to remove planktonic growth, and the tubes are rinsed twice with PBS. After washing, the tubes were stained with methylene blue for one hour [14] then rinsed twice with PBS, air-dried overnight in an inverted position, and observed for biofilm on the walls as well as bottom of the tube [14].
Virulence Genes Assessment by Multiplex PCR
Urine samples showing Gram negative rods were cultured on CLED medium and pink colonies were gram stained and biochemical tests like Indole, Methyl Red, Voges Proskauer, Citrate and motility test were done for confirmation. Multiplex PCR was done for E. coli iutA gene, papC gene, cnfl gene and hlyA gene. Invitrogen DNAzol ready-to-use reagent was used for the isolation of genomic DNA (Invitrogen™ DNAzol™ Reagent, for isolation of genomic DNA from solid and liquid samples Catalog number10503027) (Genomic DNA Isolation Reagent, Invitrogen) [15]. Primers ulmastermix@ used for a full reaction of 100 μL master mix at concentration of 100 picomoles for each primer using Invitrogen green master mix system and Invitrogen Red Taq polymerase 3U for a full reaction of 100 μL, final volume of each full reaction was made up using Ambion Nuclease free water. Thermal cycler was programmed for initial denaturation at 94°C for two minutes followed by 35 cycles of denaturation at 94°C for one minute. Primer annealing at 55°C for one minute, primer extension at 72°C for two minute and final elongation at 72°C for five minutes, at 4°C. After PCR all tubes were stored at- 20°C until electrophoresis. Agarose gel electrophoresis was done using Invitrogen ThermoFisher molecular biology reagents. Invitrogen low EEO agarose, ThermoFisher TAE buffer, Invitrogen 100 bp DNA ladder plus, ThermoFisher 6Xloading dye, HI Media EtBr400 ng/μL concentration in 2% agarose gel run for one hour at 100 volts. DNA bands were visualised on 312 nm filter followed by Gel documentation and report preparation.
Statistical Analysis
Statistical analysis was conducted using MS Excel and IBM Statistical Package for the Social Sciences (SPSS) version 27.0. Quantitative data presented as mean and SD while qualitative variables expressed as frequency (percentage).
Results
A total of 495 urine samples were screened for diagnosis of UTI, on Gram staining 149 samples observed with Gram negative bacteria. Based on morphology, 92 samples with thin gram-negative rods were processed for confocal microscopy while gram negative cocci and fungal elements were excluded. Demographics like age and clinical symptoms related to UTI are represented in [Table/Fig-2].
Demographic and symptoms in women diagnosed with UTI.
| Women diagnosed with UTI | N=92 |
|---|
| Age (years)* | 36.2±9.5 |
| Symptoms n (%) | |
| Backache | 1 (1.1) |
| Burning sensation during micturition | 19 (20.7) |
| Frequent micturition | 16 (17.4) |
| Lower abdominal pain | 11 (12.0) |
| Fever | 74 (80.4) |
| No symptoms | 45 (48.9) |
| Recurrent infection | 47 (51.1) |
Age *noted as mean (±SD), and frequencies are represented as percentages
On gram staining 149 (30.1%) samples were detected with Gram-negative rods. Based on bacterial morphology only 92 samples were studied for detection of IBC by using confocal microscopy and the remaining 57 samples were excluded because of different morphology. Detailed bacterial profile by confocal microscopy is presented in [Table/Fig-3].
Urine bacterial profile detected by confocal microscopy.
| Urine bacterial profile (n=92) | N (%) |
|---|
| IBC E. coli | 11 (12.0) |
| Extracellular E. coli | 4 (4.3) |
| Extracellular Pseudomonas | 3 (3.3) |
| Filamentous Pseudomonas | 1 (1.1) |
| IBC of Pseudomonas | 3 (3.3) |
| IIB E. coli | 23 (25.0) |
| IIB Pseudomonas | 7 (7.6%) |
| Negative for E. coli and Pseudomonas | 40 (43.5) |
IIB: Isolated intracellular bacteria; IBC: Intracellular bacterial community
Out of 92 urine samples, 23 (25%) were detected with IIB E.coli, followed by 11 (12.0%) IBC E.coli, while 1 (1.1%) sample detected with filamentous Pseudomonas and 40 (43.5%) samples were negative [Table/Fig-4]. Light microscopy using Gram and Giemsa stain were shown in [Table/Fig-5].
Urine microbial flora detected by culture method.
| Urine microbial flora (n=92) | N (%) |
|---|
| E. coli | 23 (25.0) |
| Enterococcus | 1 (1.1) |
| Klebsiella | 6 (6.5) |
| Polymicrobial flora | 29 (31.5) |
| Pseudomonas | 4 (4.3) |
| Culture negative | 29 (31.5) |
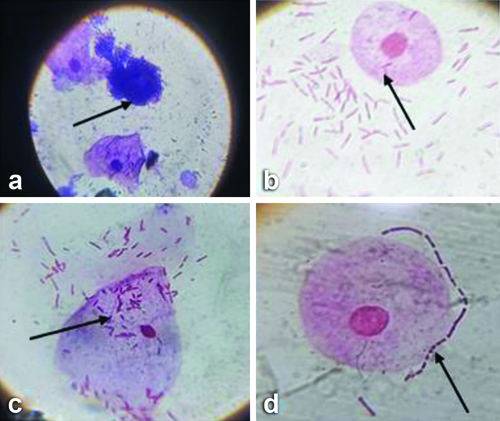
Observations under light microscopy, arrows indicating: a) Giemsa staining - darkly stained urothelial cells suggestive of IBCs; b,c) Gram staining showing IIBs; d) Showing urothelial cells surrounded by bacteria using 100x oil immersion objective that is 1000x magnification for all above images.
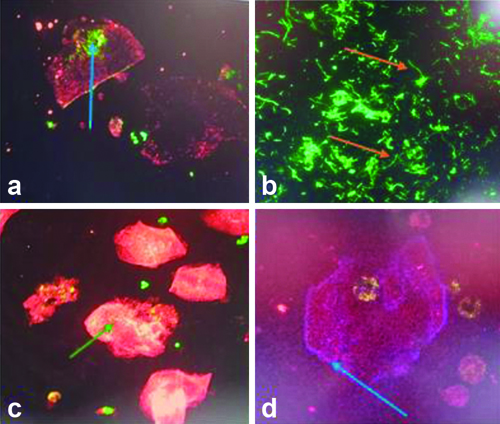
Under confocal microscopy merged images showed communities of bacteria within exfoliated urine facet cells [Table/Fig-6]. The distribution of fluorescent staining was observed with profiles of fluorescent intensity along lines drawn through the middle of IBCs. Higher peaks of uroplakin staining observed in IBCs.
Confocal Microscopy observations. a,c,d) Showing IIBs stained with FITC green, UPIII stained cytoplasm red and Alexa Fluor 350 stained cell membrane as blue; b) Showing filamentous Pseudomonas using Plan-Apochromat 63x/1.4 oil immersion objective for all the above images.
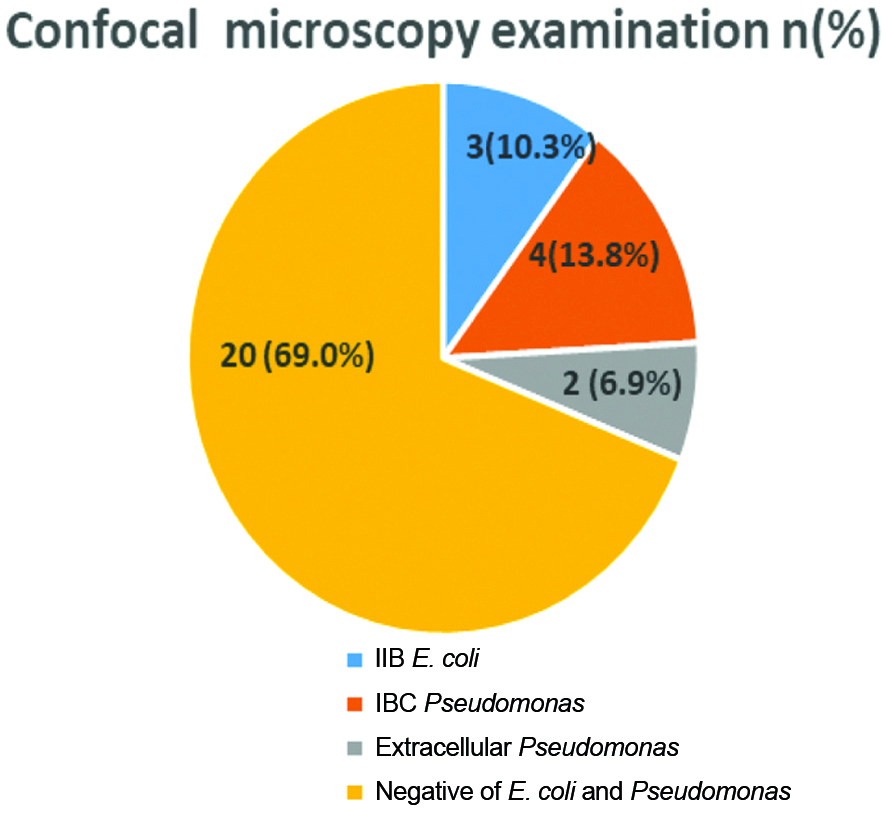
The 92 samples were processed for urine culture and the percentage of polymicrobial flora was higher 29 (31.5%) as compared to E. coli 23 (25.0%) while 29 (31.5%) urine samples were culture negative. On further analysis of 29 culture negative urine samples, nine samples were detected with IIB/IBC by confocal microscopy. UPEC strains of IIB/IBC represented in [Table/Fig-7].
Detection of IIB/IBC in culture negative urine samples (n=29).
Also, their clinical symptoms of UTI are represented in [Table Fig-8].
Clinical symptoms of UTI in women who were detected culture negative (n=29).
| Clinical symptoms | n (%) |
|---|
| Fever | 20 (69) |
| Burning sensation | 7 (24.1) |
| Backache | 1 (3.4) |
| Recurrent UTI | 8 (27.6) |
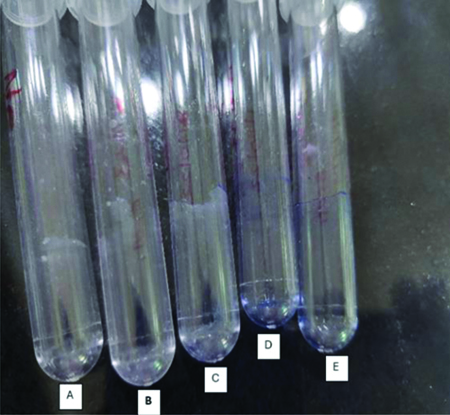
Biofilm formation was observed in nine urine samples and was confirmed using tube method [Table/Fig-9].
Biofilm detection using tube method, observed biofilm formation in tube A, B, C and other two tubes D and E are controls.
(Control tubes consist of only trypticase soya broth without any inoculation and followed by every remaining steps. We have provided a few representative images as an example for biofilm detection among all the nine test-tubes in which biofilm was detected)
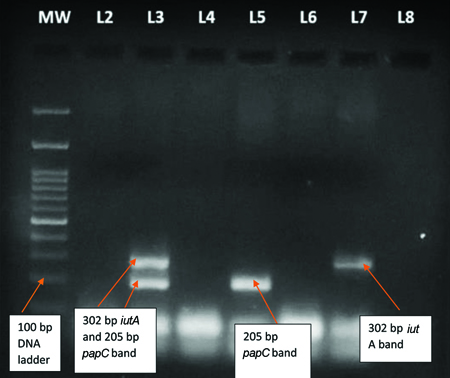
Detection of virulence gene by PCR: For the detection of UPEC, 24 urine samples were selected that were positive for E. coli by culture and confocal microscopy were processed for virulence gene detection by PCR. A total of 12 (50.0%) isolates were detected with iutA gene, 3 (12.5%) isolates detected with both iutA and papC gene, while 1 (4.1%) isolate detected with only papC [Table/Fig-10].
Multiplex PCR product observed in E. coli isolates.
MW (L1):100 bp DNA ladder plus, iutA gene with 302 bp PCR product is observed in E. coli isolates (L7), whereas both iutA and papC205 bp PCR products are observed in E. coli isolate (L3) and only papC PCR product is observed in E.coli isolate (L5)
Discussion
The UTI is one of the most common bacterial infections and more prevalent in females with incidence rate 50-fold higher among age group of 20-50 years as compared to males [16]. In India the prevalence of UTI in women of reproductive age group is documented to be 49.3% [16,17]. The majority of uncomplicated UTI are community acquired and 80-90% of them were caused by UPEC and other Gram-negative bacteria like Klebsiella and Pseudomonas [18,19], present study observed 25% E. coli and 4.3% Pseudomonas. Previously, UTI was typically considered an extracellular infection, but many human and animal studies demonstrated that IIB like E. coli and other gram-negative bacteria including Pseudomonas have the ability to invade urothelium to form IBC. Hence, IBCs are difficult to detect in urine culture. Also, easily escape from host immune mechanism and antibiotic treatment lead to recurrent UTI [16]. Most of the UTI therapeutic guidelines recommend the use of β-lactams for empirical treatments [20-22]. However, these antibiotics do not reach enough intracellular concentrations. So alternative therapy for recurrent UTI would be necessary.
IIB represents the initial stage of IBC showing small and coccoid size. While, filamentous bacteria represent the late stage of IBCs with high attachment to the urothelial cells. Such filamentous forms are difficult to phagocytise and increase bacterial resistance against strong urine forces [23,24]. However, detection of IIB/IBC is crucial in patients who are culture negative but represent clinical symptoms of UTI to avoid recurrent infection and overdosage of antibiotics. The culture negative urine samples can be diagnosed with an alternative, highly sensitive method to detect IIB/IBC. These observations emphasise the need for more sensitive techniques like confocal microscopy to detect intracellular bacteria.
There are very few research findings available on use of confocal microscopy for detection of UPEC as summarised in [Table/Fig-11] [2,3,7]. These findings demonstrated the use of confocal microscopy in conjunction with light and electron microscopy. Cheng Y et al., reported 13% bacterial cystitis on routine microbiological examination followed by Wright staining of 20 samples for confocal microscopy showed presence of 50% E. coli [2]. In present study, 29 (31.5%) urine samples were culture negative processed for confocal microscopy showed 3 (10.3%) with IIB E. coli, 4 (13.8%) with IBC Pseudomonas and 2 (6.9%) with extracellular Pseudomonas. The other study findings also showed the use of confocal microscopy as a sensitive method for the detection of IBC and supported the above findings [3,7].
Summary of literature review for studies using confocal microscopy for detection of intracellular bacteria [2,3,7].
| S. No. | Author | Study type | Study site | Techniques used | No. of samples | Results |
|---|
| 1 | Rosen DA et al., 2007 [3] | Comparative | Washington | Light, electron and confocal microscopy | 80 women with uncomplicated cystitis and 20 asymptomatic women | In urine samples with UTI, 18% (14/80) IBC, and 41% (33/80) filamentous bacteria were found. |
| 2 | Robino L et al., 2014 [7] | Observational | Uruguay | Light and confocal microscopy | 133 children | 36.8% (49/133) urine samples, the presence of IB was suspected by confocal microscopy. |
| 3 | Cheng Y et al., 2016 [2] | Case-control | Australian | Light and confocal microscopy | 88 women | On routine microbiological examination 13% (12/88) were bacterial cystitis. On confocal microscopy 50% (10/20) E. coli were detected. |
| 4. | Present study, 2025 | Observational | India | Light and confocal microscopy | 92 women | On confocal microscopy examination, 23 (25.0%) IIB and 11 (12.0%) IBC were detected.
While out of 29 culture negative samples 9 (3.1%) were detected with IIB and IBC by confocal microscopy. |
In present study, the expression of virulence genes like iutA and papC in UPEC strain was 12 (52.1%), 4 (13.7%), respectively, which was like the other studies who reported similar frequency of iutA (52%-64%) and 22% papC [7,25]. As per literature findings iutA gene was the commonest gene detected in the UPEC isolates which help bacterial iron intake and survival at limited concentration of iron in urinary tract [24,26,27]. Also, these genes are associated with antibiotic resistant stain, resistant to various antibiotics like amoxicillin-clavulanic acid, ampicillin, and cefotaxime [28-30].
In present study, virulence gene detection and biofilm formation were supported by the detection of IIB/IBC by confocal microscopy. In Indian scenario, there is a scarcity of literature on use of confocal microscopy for the detection of IIB/IBC in UTI. However, confocal microscopy has the advantage of being highly sensitive and time saver as compared to routine diagnostic methods of UTI.
Limitation(s)
The IBC may have missed out sometimes because of its transient nature. It is possible that the sensitivity of our assay might have been better if every sample was quickly processed without any delay. Serial specimens were not collected form these women and so could not optimise collection timepoints when IBCs and filaments might be most abundant. The low numbers of non-E. coli infections have made it difficult to assess the ability of other uropathogens to form filaments and IBCs during human infection. The volume of urine analysed represents a small proportion of the total sample which may underestimate the prevalence of IBC pathway. Virulence genes have not been assessed in isolated Pseudomonas because of budget restriction.
Conclusion(s)
The diagnosis of IIB/IBC is crucial in suspected UTI where routine urine culture methods fail to detect the bacteria. Present study findings suggested that detection of intracellular bacteria using confocal microscopy along with virulence gene assessment played a significant role in understanding the pathophysiology of UTIs. Hence, the use of confocal microscopy with diagnostic approach may serve as a better option for identification and management of IIB/IBC in patients with recurrent UTI or clinical symptoms with negative culture findings.
Age *noted as mean (±SD), and frequencies are represented as percentages
IIB: Isolated intracellular bacteria; IBC: Intracellular bacterial community