Introduction
The widespread use of antibiotics has been pivotal in controlling bacterial infections; yet, it has also led to the rapid emergence of antibiotic-resistant microorganisms [1]. This phenomenon presents a significant challenge to global health, as it threatens the effectiveness of current treatments. When exposed to antibiotic stress, microbes employ a variety of mechanisms to survive and proliferate, often developing resistance through genetic mutations, HGT and adaptive changes in their physiology [2]. These adaptations not only enable individual survival but also promote the persistence of resistant strains within microbial communities. Understanding the intricacies of microbial responses to antibiotic pressure is critical for developing novel approaches to combat resistance and ensure the continued efficacy of antimicrobial agents [3,4]. This review aims to delve into the mechanisms by which microbes respond to antibiotic stress, highlighting both well-established and emerging strategies of resistance and adaptation.
Literature Search
A narrative review was conducted on microbial responses to antibiotic stress. The focus was to identify and analyse key mechanisms of resistance, including genetic and phenotypic adaptations, such as HGT, biofilm formation and mutations. The review incorporated both quantitative and qualitative data from various sources, including peer-reviewed articles, experimental studies and relevant clinical research. The search for relevant literature on antibiotic resistance was conducted using academic databases such as PubMed, Scopus and Google Scholar. The search strategy incorporated the following keywords: “antibiotic resistance,” “microbial resistance mechanisms,” “HGT”, “biofilm formation,” “mutations in bacteria,” “efflux pumps,” and “adaptive dosing strategies.” Boolean operators (AND, OR) were applied to refine and expand the search results, ensuring a comprehensive capture of relevant articles. The combination of keywords was used in various configurations, including: “antibiotic resistance” AND “microbial resistance mechanisms” “HGT” OR “biofilm formation”, “mutations in bacteria” AND “efflux pumps” “adaptive dosing strategies” AND “antibiotic resistance”.
These combinations helped explore different aspects of microbial resistance and adaptive mechanisms in response to antibiotic pressure, facilitating a broad and deep understanding of the topic.
Inclusion criteria: Peer-reviewed studies published between 2000 and 2024. Research on both genetic and phenotypic mechanisms of bacterial resistance to antibiotics. Articles providing insights into innovative antimicrobial strategies, such as multitarget drugs and adaptive dosing were included in the study.
Exclusion criteria: Studies focusing on viral or fungal resistance. Research not including bacterial mechanisms of antibiotic resistance were excluded from the study.
Primary data were obtained from laboratory studies involving in-vitro bacterial cultures to observe microbial resistance development under different antibiotic pressures. Techniques such as whole-genome sequencing, Polymerase Chain Reaction (PCR) and bioinformatics were employed to assess genetic adaptations. Additionally, phenotypic responses, such as the activation of efflux pumps and changes in cell wall permeability, were evaluated through biochemical assays and microscopy.
Mathematical modeling, particularly using MATLAB (matrix laboratory), was used to simulate resistance emergence, mutation distributions and the effects of adaptive dosing strategies. These models provided insight into the interactions between bacterial populations and antibiotic treatments, contributing to predictions regarding the efficacy of novel antimicrobial therapies.
Genetic Mechanisms Underlying Antibiotic Resistance
Genetic mechanisms play a crucial role in the development and spread of antibiotic resistance among microbial populations. One of the primary methods through which bacteria acquire resistance is HGT [5]. This process allows bacteria to exchange genetic material, including resistance genes, through mechanisms like conjugation, transformation and transduction. As a result, resistance can rapidly spread within bacterial species, particularly in environments with high antibiotic usage, such as hospitals and agricultural settings [6]. The widespread presence of plasmids, transposons and integrons in bacteria facilitates this exchange, making HGT a significant factor in the proliferation of antibiotic resistance. In addition to HGT, spontaneous mutations in bacterial DNA can also lead to resistance [Table/Fig-1] [7].
Mutation frequency distribution.
*Type 1: Point mutation- Small changes in DNA, leading to amino acid substitutions; *Type 2: Insertions- Addition of nucleotides, which can disrupt or enhance gene function; *Type 3: Deletions- Removal of nucleotides, potentially causing gene loss; *Type 4: Duplications- Extra copies of DNA segments, increasing gene dosage; *Type 5: Inversions- DNA segments flipped, affecting gene regulation; *Type 6: Translocations- DNA moved to new locations, disrupting gene function; *Type 7: Large Deletions/Rearrangements- Major DNA changes, impacting multiple genes or operons
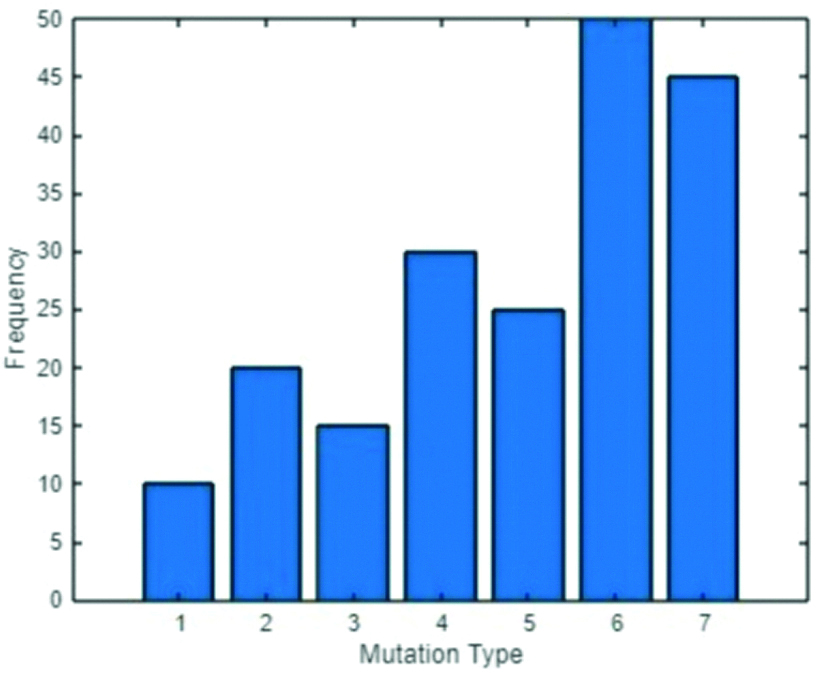
For mutation frequency evaluation in a bacterial population exposed to antibiotics, a systematic approach was utilised. Initially, bacteria are subjected to controlled laboratory conditions with antibiotics, from which individual colonies are isolated for further analysis. Following isolation, genomic sequencing is performed on the DNA extracted from these isolates to identify the specific mutations present. This data is then analysed using bioinformatics tools to gain insights into the genetic changes. In the Mutation Frequency Distribution plot [Table/Fig-1], the x-axis represents different mutations observed in a bacterial population subjected to antibiotic exposure. Each mutation type, labeled from 1 to 7, corresponds to specific genetic changes that can confer varying degrees of resistance. Each unique mutation is counted to determine its frequency within the population, expressed as a percentage of the total number of mutations identified across all samples. The bar graph is plotted by representing the frequency of each mutation type on the y-axis, typically expressed in percentages (n %). Each bar’s height indicates the relative abundance of that specific mutation type within the population.
These mutations often occur in genes that encode the target sites of antibiotics, such as the “gyrA” gene involved in quinolone resistance or ribosomal RNA genes that confer resistance to macrolides and aminoglycosides. When these mutations alter the antibiotic’s binding site, they can prevent the drug from effectively interacting with its target, rendering the treatment ineffective [Table/Fig-2] [8]. The effect of mutations on the antibiotic binding affinity plot [Table/Fig-2] illustrates how specific genetic mutations influence the binding capability of various antibiotics to their bacterial targets. The x-axis categorises different types of genetic mutations, such as point mutations, insertions, deletions and duplications, which occur in genes related to antibiotic resistance. The y-axis represents the binding affinity of specific antibiotics, such as penicillin, macrolides, and fluoroquinolones. This plot demonstrates that certain mutations can lead to a significant reduction in the binding affinity of these antibiotics, thereby diminishing their effectiveness in inhibiting bacterial growth, as shown in [Table/Fig-2]. As binding affinity decreases, the bacteria develop increased resistance to the antibiotic, making it less effective as a treatment option. This visualisation is crucial for understanding the molecular basis of antibiotic resistance, as it directly links specific genetic alterations to functional outcomes in bacterial survival.
Effect of mutations on the binding affinity of antibiotics.
*X-Axis (Mutation Types): 1: Point Mutation- A single nucleotide change, potentially altering an amino acid in a key protein; 1.5: Insertion- Addition of a small number of nucleotides that may disrupt gene function; 2: Deletion- Removal of nucleotides, potentially causing loss of protein function; 2.5: Duplication- Repetition of a DNA segment, increasing gene dosage; 3: Inversion- Reversal of a DNA segment, affecting gene regulation; 3.5: Translocation- Relocation of DNA segments, which can disrupt essential genes; 4: Large deletion- A significant deletion that impacts multiple genes or operons; 4.5: Large Rearrangement- Major structural alteration that affects gene clusters.
*Y-Axis (Binding Affinity Values): 0.8: High binding affinity, indicating strong antibiotic-target interaction; 0.7: Moderate binding affinity, with slightly reduced antibiotic efficacy; 0.6: Noticeable decrease in binding affinity, reducing antibiotic effectiveness; 0.5: Significant reduction in binding affinity, limiting the drug’s impact on the bacteria; 0.3: Very low binding affinity, meaning the antibiotic is barely effective; 0.2: Minimal binding affinity, essentially rendering the antibiotic ineffective
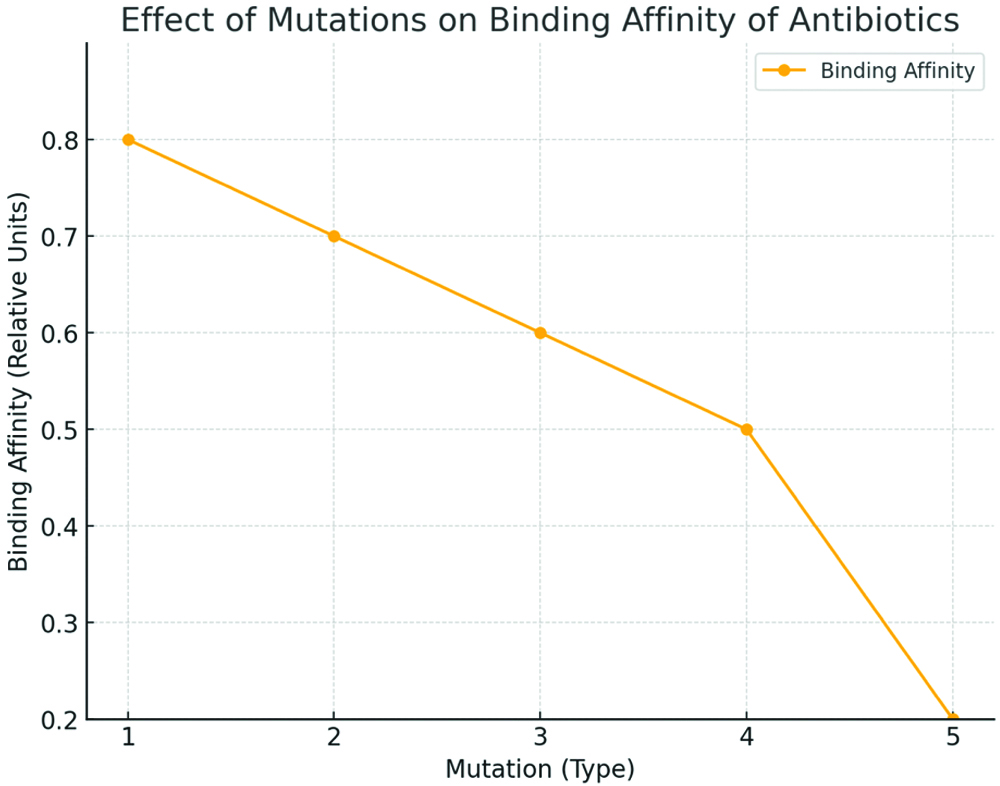
By clearly depicting how mutations can alter drug-target interactions, the plot underscores the importance of considering these molecular changes when developing new antibiotics or modifying existing ones. Furthermore, it highlights the challenges in overcoming resistance, as even minor genetic changes can profoundly affect the efficacy of antimicrobial treatments. Understanding these interactions is essential for designing strategies aimed at restoring the binding affinity of existing drugs or developing new compounds that effectively target resistant strains.
The selective pressure exerted by antibiotics ensures that bacteria with such beneficial mutations survive and proliferate, further contributing to the resistance problem. The combined effects of HGT and spontaneous mutations create a dynamic and challenging landscape for antibiotic treatment. As bacteria continue to adapt genetically, the efficacy of existing antibiotics diminishes, leading to the emergence of multidrug-resistant strains [Table/Fig-3] [9].
Resistance emergence simulation plot.
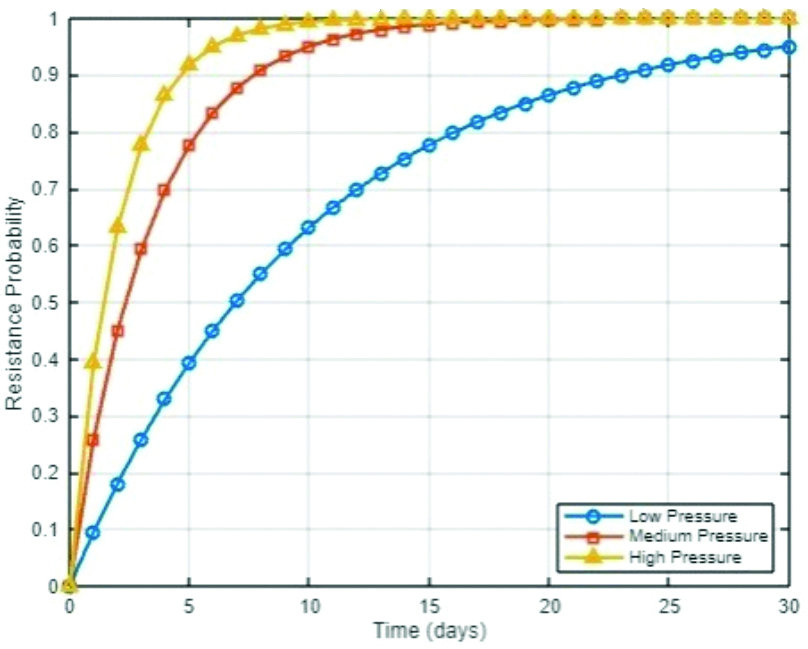
The resistance emergence simulation plot provides insights into how different levels of antibiotic selective pressure can influence the speed at which bacterial resistance emerges. By modeling resistance emergence as a function of selective pressure, the plot reveals that higher selective pressures, such as those exerted by more potent or continuously administered antibiotics, can accelerate the development of resistance [10]. This is particularly evident in the steep rise of resistance probability over time in scenarios where antibiotics are used aggressively. The plot serves as a visual representation of the delicate balance between effectively killing bacteria and inadvertently promoting resistance through excessive or inappropriate antibiotic use.
Understanding the genetic mechanisms behind resistance is essential for developing new strategies to combat bacterial infections and for designing next-generation antibiotics that can overcome these adaptive responses. In addition to the direct genetic changes that confer antibiotic resistance, bacteria can also evolve more complex mechanisms that enhance their survival under antibiotic pressure. Over time, bacteria may accumulate [11].
Multiple resistance genes are often carried on mobile genetic elements such as plasmids, which can lead to the development of multidrug-resistant strains [Table/Fig-4]. These strains are particularly concerning because they can resist various antibiotics, making infections increasingly difficult to treat [12]. The persistence and spread of these multidrug-resistant bacteria are exacerbated by environments with constant antibiotic exposure, which continually select for the most resistant strains. To effectively combat this growing threat, it is crucial to understand the genetic evolution of resistance and to focus on developing new antibiotics and strategies that can outpace these adaptive changes [13].
Multitarget drug efficacy plot.
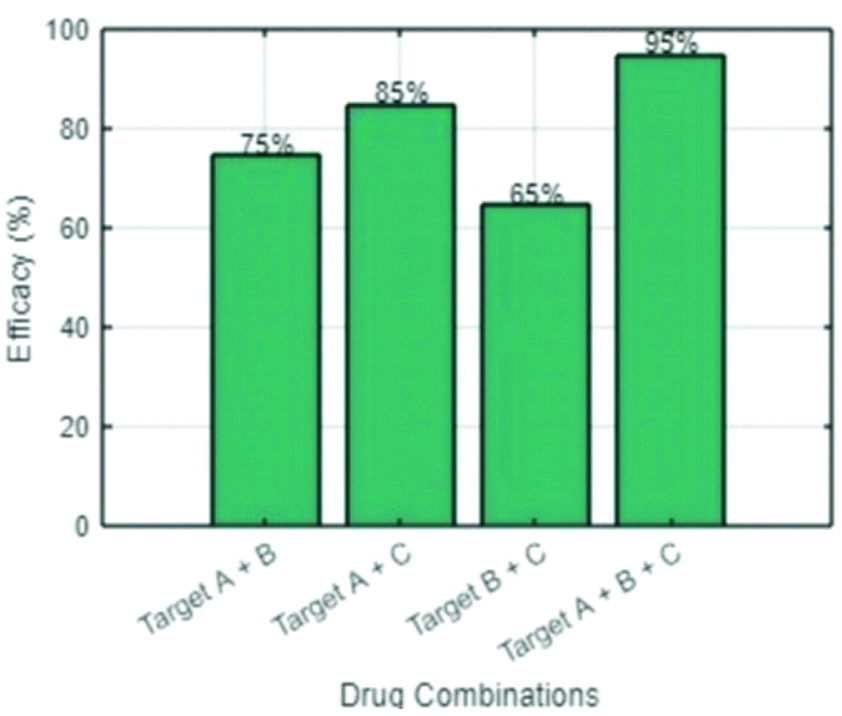
Efficacy is calculated by comparing bacterial growth reductions in treatment groups relative to the control, often expressed as percent inhibition or Minimum Inhibitory Concentration (MIC). In the efficacy study, Group A is one of the treatment groups that is provided with the main experimental intervention, such as a new multitarget antibiotic or a combination of drugs targeting multiple pathways within bacteria. Group B would be another treatment group with an alternative approach: either a different drug, varying drug combinations, or less aggressive regimens. This way, researchers can compare the outcomes of Groups A and B to see which treatment is more effective in reducing bacterial growth. Group C, which is the control group, serves as the baseline, receiving either a placebo or standard treatment that does not target multiple pathways. This helps establish the natural progression of bacterial growth without the influence of new or alternative treatments. By measuring bacterial inhibition within the groups, typically using percentage inhibition that denotes the decrease in bacterial growth compared to the control, or MIC, which is the lowest concentration of a treatment preventing evident growth of bacteria, the relative efficacies of interventions used in each group are determined. This setup sheds light on the best treatment methodologies to ensure that bacterial growth does not increase and that the causative bacteria do not develop any resistance. This approach leverages the principle that bacteria would need to undergo multiple simultaneous changes to evade the effects of a multitarget drug, making resistance less likely to develop. The plot demonstrates that certain combinations, particularly those that target more than two pathways, result in a substantial reduction in bacterial growth, as shown by the higher efficacy percentages.
The HGT network plot offers a detailed visualisation of the genetic exchange processes that contribute to the spread of antibiotic resistance among bacterial species. Each node within this network represents a distinct bacterial species and the edges connecting these nodes indicate the transfer of antibiotic-resistance genes. The thickness of the edges is directly proportional to the frequency of these gene transfer events, revealing how often resistance genes are shared between species. This visual representation emphasises the significance of HGT as a primary mechanism through which bacteria acquire resistance, allowing them to survive in environments with high antibiotic usage. The ability of bacteria to exchange genetic material so readily through processes like conjugation, transformation and transduction makes HGT a powerful force in the rapid dissemination of resistance traits across diverse microbial populations [14].
The interconnected nature of the HGT network illustrates the complexity of gene flow within microbial communities [15]. Unlike mutations within individual organisms, HGT allows for the horizontal movement of genes across different species, often crossing traditional species boundaries. This means that resistance genes can quickly spread from one bacterial population to another, even if the species are not closely related. The network visualisation highlights how resistance genes can be shared within a local microbial community, creating a reservoir of resistance traits that can be accessed by different species as needed. This not only accelerates the spread of resistance but also complicates efforts to control it, as resistance can easily move between species that inhabit the same environment, such as within hospitals or agricultural settings [16]. Moreover, the HGT network plot underscores the challenge of combating antibiotic resistance in the context of these gene transfer dynamics.
Phenotypic Adaptations and Survival Strategies
Phenotypic adaptations are essential mechanisms that bacteria employ to survive under antibiotic stress. Unlike genetic changes, which involve alterations in the DNA sequence, phenotypic adaptations allow bacteria to adjust their behaviour and physiology in response to environmental pressures without changing their genetic code. Key mechanisms include changes in cell wall permeability, which restrict antibiotic entry; activation of efflux pumps that expel antibiotics, reducing their intracellular concentration; and alterations in metabolic pathways that allow bacteria to utilise alternative nutrients. Additionally, biofilm formation provides a protective barrier against antibiotics, enhancing resistance by preventing drug penetration. Phenotypic heterogeneity within bacterial populations means some cells may exhibit greater resistance, contributing to overall survival during treatment. Finally, bacteria activate stress response mechanisms to repair damage and mitigate antibiotic effects. Understanding these adaptations is crucial for developing effective therapeutic strategies against bacterial resistance [17].
One key phenotypic adaptation is the modification of cell wall structure. When exposed to antibiotics that target cell wall synthesis, some bacteria can alter their cell wall composition to reduce the effectiveness of the drug. For example, by thickening the cell wall or modifying its chemical composition, bacteria can prevent antibiotics from penetrating the cell or reaching their targets. This strategy is particularly common in gram-positive bacteria, which possess a thick peptidoglycan layer that can be adjusted to block antibiotic entry [18,19].
Efflux pumps represent another crucial survival strategy. These are protein complexes that actively transport antibiotics out of the bacterial cell, reducing the intracellular concentration of the drug to sublethal levels. By upregulating the expression of efflux pumps, bacteria can effectively decrease the impact of antibiotics, allowing them to survive in hostile environments. MATLAB can be used to model the kinetics of efflux pump activity, illustrating how changes in pump efficiency affect bacterial survival under different antibiotic concentrations [20,21].
Implications for Future Antimicrobial Development and Strategies
The ongoing rise of antibiotic resistance necessitates innovative approaches to antimicrobial development and deployment strategies. As bacteria continue to adapt and develop resistance through both genetic mutations and phenotypic adaptations, the effectiveness of existing antibiotics is diminishing. This creates an urgent need for new antimicrobials that can outpace bacterial evolution. Future strategies must focus not only on the development of novel drugs but also on improving the effectiveness of existing treatments and implementing smarter usage practices to extend the lifespan of current antibiotics.
One critical aspect of future antimicrobial development is the design of drugs that target multiple bacterial processes simultaneously. By attacking several pathways or structures within bacteria, such as cell wall synthesis, protein production, and DNA replication, these drugs reduce the likelihood of resistance developing through single mutations or adaptations. MATLAB can be used to simulate the effectiveness of these multitarget drugs, providing insights into how different combinations of targets can inhibit bacterial growth and reduce the emergence of resistance.
In addition to new drug development, optimising the dosing strategies of existing antibiotics is crucial. Adaptive dosing strategies, which adjust drug administration based on real-time monitoring of bacterial response, could minimise the selective pressure that drives resistance [Table/Fig-5]. These models could explore the balance between drug efficacy and the potential for resistance, guiding the design of treatments that maintain therapeutic effectiveness while reducing the risk of resistance [22,23].
Adaptive dosing strategy plot.
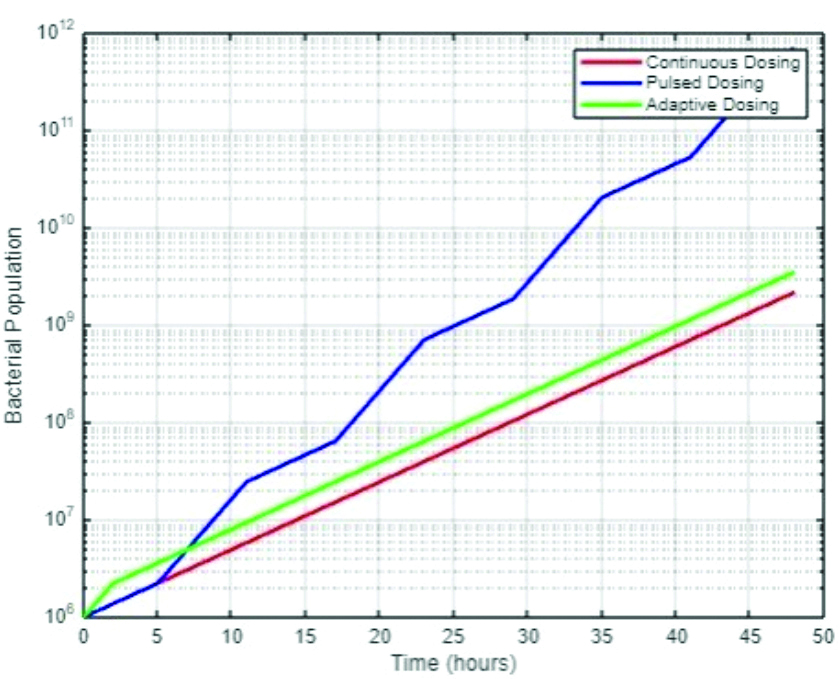
This simulation highlights the critical need for carefully calibrated antibiotic use in both clinical and environmental settings. The data suggest that while strong selective pressure may initially appear to be effective, it can lead to the rapid emergence of resistant strains, ultimately undermining the long-term success of treatment [24].
The adaptive dosing strategy plot explores the potential benefits of adjusting antibiotic administration based on real-time bacterial population dynamics. Unlike continuous or pulsed dosing, which applies a consistent or predetermined antibiotic regimen, adaptive dosing strategies respond to fluctuations in bacterial populations, increasing antibiotic levels when populations exceed a certain threshold and reducing them when populations are under control [Table/Fig-5]. The plot demonstrates that adaptive dosing can more effectively manage bacterial populations over time, preventing the spikes in population seen with other strategies and potentially reducing the selective pressure that drives resistance.
Additionally, this approach offers a promising strategy for extending the useful lifespan of antibiotics by optimising their use according to the immediate needs of the patient’s condition. By avoiding unnecessary exposure to high antibiotic levels, adaptive dosing could mitigate the selective pressures that foster the development of resistant bacteria [24].
Conclusion(s)
The microbial response to antibiotic stress is a complex and multifaceted process that involves both genetic and phenotypic adaptations, which together contribute to the development and persistence of antibiotic resistance. The exploration of genetic mechanisms, including HGT and spontaneous mutations, highlights how microbes can rapidly acquire and propagate resistance traits, making them formidable opponents in the fight against infections. The analysis of phenotypic adaptations, such as biofilm formation, altered metabolic pathways and the activation of stress response systems, further underscores the ability of microbes to thrive even in hostile, antibiotic-rich environments.
The implications of these resistance mechanisms for future antimicrobial development are profound. The multitarget drug efficacy plot demonstrates the potential of innovative drug combinations to overcome resistance by targeting multiple bacterial pathways simultaneously, thereby reducing the likelihood of resistance emergence. The resistance emergence simulation plot illustrates the critical importance of balancing antibiotic potency with the risk of resistance development, emphasising the need for carefully calibrated dosing strategies. Finally, the adaptive dosing strategy plot suggests that dynamic, responsive antibiotic regimens could offer a more effective approach in managing bacterial populations while minimising selective pressures that drive resistance.
Overall, this paper underscores the urgent need for new antimicrobial strategies that are informed by a deep understanding of microbial resistance mechanisms. By integrating genetic insights with advanced dosing strategies and multitarget approaches, the development of next-generation antibiotics can be better aligned with the evolving landscape of microbial resistance, ensuring that effective treatment options remain available in the future.
[1]. Demain AL, Antibiotics: Natural products essential to human healthMed Res Rev 2009 29(6):821-42.10.1002/med.2015419291695 [Google Scholar] [CrossRef] [PubMed]
[2]. Walsh C, Molecular mechanisms that confer antibacterial drug resistanceNature 2000 406(6797):775-81.10.1038/3502121910963607 [Google Scholar] [CrossRef] [PubMed]
[3]. Konwar AN, Hazarika SN, Bharadwaj P, Thakur D, Emerging non-traditional approaches to combat antibiotic resistanceCurr Microbiol 2022 79(11):33010.1007/s00284-022-03029-736155858PMC9510247 [Google Scholar] [CrossRef] [PubMed]
[4]. Davies J, Davies D, Origins and evolution of antibiotic resistanceMicrobiol Mol Biol Rev 2010 74(3):417-33.10.1128/MMBR.00016-1020805405PMC2937522 [Google Scholar] [CrossRef] [PubMed]
[5]. Bello-López JM, Cabrero-Martínez OA, Ibáñez-Cervantes G, Hernández-Cortez C, Pelcastre-Rodríguez LI, Gonzalez-Avila LU, Horizontal gene transfer and its association with antibiotic resistance in the genus Aeromonas sppMicroorganisms 2019 7(9):36310.3390/microorganisms709036331540466PMC6780555 [Google Scholar] [CrossRef] [PubMed]
[6]. Džidić S, Šušković J, Kos B, Antibiotic resistance mechanisms in bacteria: biochemical and genetic aspects. Antibiotic Resistance in Bacteria, Food TechnolBiotechnol 2008 46(1):11-21. [Google Scholar]
[7]. Taylor PL, Gilman CF, Blaskovich HA, Antibiotic resistance: Mechanisms and new strategies to combat an ancient problemScience 2018 360(6390):1110-15. [Google Scholar]
[8]. Munita JM, Arias CA, Mechanisms of antibiotic resistanceMicrobiol Spectr 2016 4(2):VMBF-0016-201510.1128/microbiolspec.VMBF-0016-201527227291PMC4888801 [Google Scholar] [CrossRef] [PubMed]
[9]. Ali J, Rafiq QA, Ratcliffe E, Antimicrobial resistance mechanisms and potential synthetic treatmentsFuture Sci OA 2018 4(4):FSO29010.4155/fsoa-2017-010929682325PMC5905577 [Google Scholar] [CrossRef] [PubMed]
[10]. Lupo A, Coyne S, Berendonk TU, Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodiesFront Microbiol 2012 3:1810.3389/fmicb.2012.0001822303296PMC3266646 [Google Scholar] [CrossRef] [PubMed]
[11]. Reygaert WC, An overview of the antimicrobial resistance mechanisms of bacteriaAIMS Microbiol 2018 4(3):482-501.10.3934/microbiol.2018.3.48231294229PMC6604941 [Google Scholar] [CrossRef] [PubMed]
[12]. Partridge SR, Kwong SM, Firth N, Jensen SO, Mobile genetic elements associated with antimicrobial resistanceClin Microbiol Rev 2018 31:00088-17.Available from: https://doi.org/10.1128/cmr.00088-1710.1128/cmr.00088-1730068738PMC6148190 [Google Scholar] [CrossRef] [PubMed]
[13]. Bharadwaj A, Rastogi A, Pandey S, Gupta S, Sohal JS, Multidrug-resistant bacteria: Their mechanism of action and prophylaxisBiomed Res Int 2022 2022:541987410.1155/2022/541987436105930PMC9467707 [Google Scholar] [CrossRef] [PubMed]
[14]. Zhou H, Beltrán JF, Brito IL, Functions predict horizontal gene transfer and the emergence of antibiotic resistanceSci Adv 2021 7(43):eabj5056Epub 2021 Oct 2210.1126/sciadv.abj505634678056PMC8535800 [Google Scholar] [CrossRef] [PubMed]
[15]. McInnes RS, McCallum GE, Lamberte LE, van Schaik W, Horizontal transfer of antibiotic resistance genes in the human gut microbiomeCurr Opin Microbiol 2020 53:35-43.Available from: https://doi.org/10.1016/j.mib.2020.02.00210.1016/j.mib.2020.02.00232143027 [Google Scholar] [CrossRef] [PubMed]
[16]. Jain R, Rivera MC, Moore JE, Lake JA, Horizontal gene transfer accelerates genome innovation and evolutionMolecular Biology and Evolution 2003 20(10):1598-602.Available from: https://doi.org/10.1093/molbev/msg15410.1093/molbev/msg15412777514 [Google Scholar] [CrossRef] [PubMed]
[17]. Spratt MR, Lane K, Navigating environmental transitions: The role of phenotypic variation in bacterial responsesmBio 2022 13(6):e0221222Epub 2022 Oct 1910.1128/mbio.02212-2236259726PMC9765552 [Google Scholar] [CrossRef] [PubMed]
[18]. Wee GN, Lyou ES, Hong JK, No JH, Kim SB, Lee TK, Phenotypic convergence of bacterial adaption to sub-lethal antibiotic treatmentFront Cell Infect Microbiol 2022 12:91341510.3389/fcimb.2022.91341536467735PMC9714565 [Google Scholar] [CrossRef] [PubMed]
[19]. Parvin F, Rahman MA, Deva AK, Vickery K, Hu H, Staphylococcus aureus cell wall phenotypic changes associated with biofilm maturation and water availability: A key contributing factor for chlorine resistanceInt J Mol Sci 2023 24(5):498310.3390/ijms2405498336902413PMC10003762 [Google Scholar] [CrossRef] [PubMed]
[20]. Gaurav A, Bakht P, Saini M, Pandey S, Pathania R, Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitorsMicrobiology (Reading) 2023 169(5):00133310.1099/mic.0.00133337224055PMC10268834 [Google Scholar] [CrossRef] [PubMed]
[21]. Reens AL, Crooks AL, Su CC, Nagy TA, Reens DL, Podoll JD, A cell-based infection assay identifies efflux pump modulators that reduce bacterial intracellular loadPLoS Pathog 2018 14(6):e100711510.1371/journal.ppat.100711529879224PMC6007937 [Google Scholar] [CrossRef] [PubMed]
[22]. Karam G, Chastre J, Wilcox MH, Vincent JL, Antibiotic strategies in the era of multidrug resistanceCrit Care 2016 20(1):13610.1186/s13054-016-1320-727329228PMC4916531 [Google Scholar] [CrossRef] [PubMed]
[23]. Rawson TM, Wilson RC, O’Hare D, Herrero P, Kambugu A, Lamorde M, Optimizing antimicrobial use: Challenges, advances and opportunitiesNat Rev Microbiol 2021 19(12):747-58.Epub 2021 Jun 2210.1038/s41579-021-00578-934158654 [Google Scholar] [CrossRef] [PubMed]
[24]. Muteeb G, Rehman MT, Shahwan M, Aatif M, Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative reviewPharmaceuticals (Basel) 2023 16(11):161510.3390/ph1611161538004480PMC10675245 [Google Scholar] [CrossRef] [PubMed]