Introdcution
Breast cancer represents a significant global health challenge, with rising incidence rates worldwide, including India, where it is a major cause of cancer-related mortality among women [1,2]. Traditionally, breast cancer prognosis and therapeutic decisions were guided by histopathological assessments, such as tumour grade and hormone receptor status. However, advancements in molecular genetics over the past two decades have led to an increased focus on identifying genetic mutations that drive breast cancer’s complex biology of breast cancer. This shift has enabled more precise diagnostic and therapeutic approaches, particularly through the identification of specific mutations associated with aggressive disease characteristics [3,4].
Since genetic mutations impair vital cell communication and cell cycle processes, encourage unchecked cell proliferation, their function in the pathophysiology of breast cancer is becoming better understood. The BRCA1 and BRCA2 gene mutations, which are known to be linked to an increased risk of hereditary breast cancer, were the focus of early research. Other genes linked to different clinical outcomes in breast cancer, including TP53, STK11, and CDH1, have been found in isolated cases [5-7]. These results highlight how crucial it is to comprehend the molecular changes that underlie the heterogeneity and progression of breast cancer.
The function of the BRAFV600E mutation in the BRAF gene is a topic of growing study. Cell division, differentiation and death are all regulated by the RAF/MEK/ERK signaling cascade, a protein encoded by BRAF and part of the Mitogen-Activated Protein Kinase (MAPK) pathway. The BRAFV600E mutation, which replaces valine at codon 600 with glutamic acid, results in constitutive BRAF protein activation. This mutation activates the MAPK pathway continuously, leading to tumourigenesis and unchecked cell proliferation [8,9]. Although melanoma is the most common type associated with BRAF, it has also been linked to thyroid and colorectal malignancies. However, its prevalence and significance in breast cancer, especially in IDC, remains unclear [10-12].
Understanding the possible significance of BRAFV600E subtypes of breast cancer, such as HER2 (human epidermal growth factor receptor 2)-negative tumours and TNBC, has been the main emphasis. Research indicates that although BRAFV600E is less common in breast cancer than in melanoma, it may exist in aggressive subgroups like TNBC and may be associated with unfavourable clinicopathological traits, such as hormone receptor negativity and higher tumour grade. This association has potential therapeutic implications since BRAF inhibitors, effective in BRAF-mutant melanoma, have shown limited efficacy in breast cancer, prompting further studies on combined BRAF and MEK inhibition strategies [13-16].
PCR techniques have been used in BRAFV600E research in breast cancer to find mutations in tumour samples. Its clinical significance in IDC is unclear and there is still a great deal of variation in the reported prevalence of BRAFV600E across various breast cancer studies [7,13,14]. Although there is insufficient information to draw firm conclusions, preliminary research indicates that BRAFV600E may be associated with aggressive disease behaviour and could contribute to treatment resistance in particular IDC subtypes [7,13,14]. This demonstrates the potential value of BRAFV600E as a therapeutic target and prognostic marker, indicating the need for additional research to elucidate its function in breast cancer [9,15].
Studies on BRAFV600E in breast cancer are rare in India, while the disease is very common. This indicates a substantial information gap about the incidence of BRAFV600E and its clinicopathological correlations in the Indian population. Understanding the frequency and significance of BRAFV600E in Indian women could provide crucial information for creating more individualised and successful treatment plans, especially considering the country’s genetic variety and distinct environmental effects. Examining this mutation may help develop customised strategies that cater to the unique requirements of Indian breast cancer patients, which could direct future treatment choices and enhance patient outcomes [2,17-19].
Hence, this narrative review summarises the body of research on PCR-based BRAFV600E mutational analysis in breast cancer, with a focus on IDC cases. It seeks to assess the frequency of the mutation, investigate correlations with clinicopathological traits, and pinpoint current research gaps. This review aims to elucidate the mutation’s potential as a prognostic and therapeutic marker by concentrating on both global and Indian contexts. This will improve clinical understanding and management of breast cancer across a variety of patient populations.
BRAFV600E MUTATION: STRUCTURE AND FUNCTIONAL IMPLICATIONS
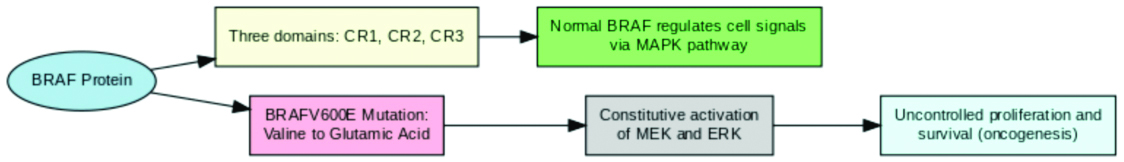
An essential component of the RAF kinase family, the BRAF protein regulates and signals cells via the MAPK pathway. In its normal state, BRAF is a serine/threonine-protein kinase that transduces extracellular signals through the RAS-RAF-MEK-ERK cascade, which regulates critical homeostasis-related activities such as cell division, differentiation and survival [5,8]. BRAF consists of three structurally conserved sections: CR1 for binding to RAS, CR2 kinase domain CR3 which is necessary for the protein’s enzymatic activity [3,5]. BRAF is normally dormant until activated by RAS, which causes MEK and ERK, promoting cell cycle progression and growth when necessary.
The V600E mutation in the BRAF gene, which replaces valine with glutamic acid at codon 600, drastically changes how BRAF functions. This mutation disrupts the kinase domain, leading to constitutive activation of BRAF even in the absence of upstream RAS signals [3,5]. Consequently, the mutant BRAF protein continuously activates MEK and ERK, resulting in unchecked cell proliferation and survival, a common occurrence in neoplastic processes [4,5]. The BRAFV600E mutation thus represents a potent oncogenic driver, found frequently in cancers such as melanoma, where it contributes to aggressive tumour growth and resistance to standard therapies [5,10,20]. This is well illustrated in [Table/Fig-1].
Flowchart illustrating BRAFV600E mutation, structure and oncogenic functional implications.
Signaling pathways involved in BRAFV600E: The MAPK/ERK pathway, a crucial signaling axis in cell control, is the main target of the BRAFV600E mutation. Extracellular cues typically cause RAS to become activated, which in turn sets off BRAF, MEK and ERK in that order. To maintain strict control over cell activity, this pathway controls the expression of genes linked to differentiation, apoptosis and proliferation [5,9]. However, BRAFV600E circumvents natural regulatory checkpoints and keeps the route open all the time [3,8]. This deregulation enhances cellular proliferation and survival, often contributing to cancer initiation, progression and metastasis in several malignancies, including breast cancer [Table/Fig-2] [8,9,11].
Flowchart depicting BRAFV600E-driven signaling pathways and downstream oncogenic effects.
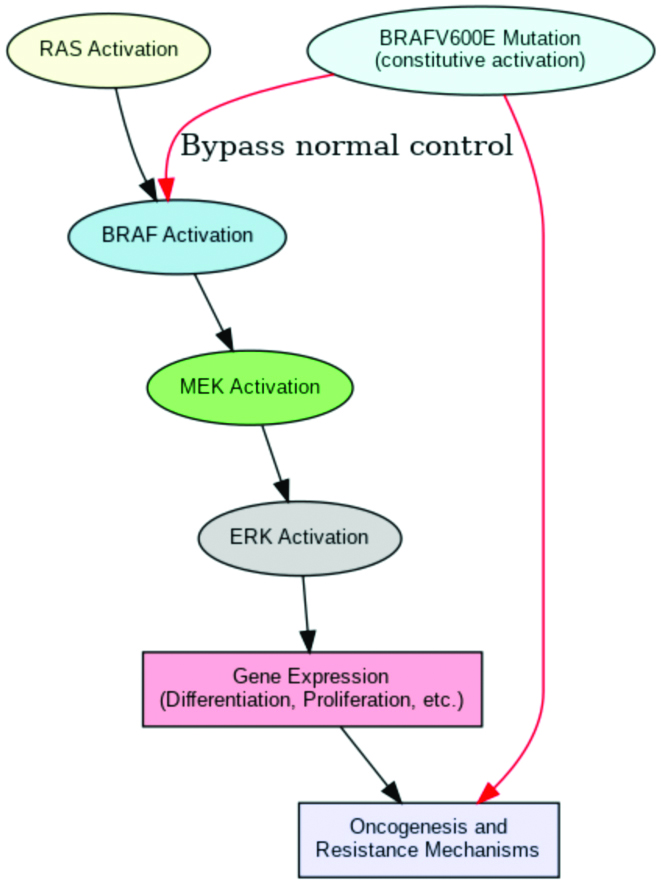
The abnormal activation of MAPK/ERK due to BRAFV600E also leads to cross-talk with other pathways, such as the PI3K/AKT pathway, further promoting oncogenesis. These interconnected pathways facilitate the cancer cells’ resilience to stress and treatment agents by intensifying signals for growth and survival [7,18,19]. The therapeutic importance of targeting BRAFV600E is highlighted by the fact that abnormalities in this pathway are directly linked to resistance to hormone therapy and other targeted treatments in breast cancer [14,21].
Impact of BRAFV600E on cancer biology: The carcinogenic potential of BRAFV600E has been thoroughly investigated in malignancies such as thyroid, colorectal and melanoma, where it plays a major role in aggressive development patterns and poor prognosis [5,10,15]. Targeted BRAF inhibitors are frequently required as part of the therapy plan for melanoma, since BRAFV600E is a significant driving mutation that promotes tumour survival and immune evasion [10,15,20]. Emerging research supports the use of combination BRAF and MEK inhibition techniques in colorectal cancer, where BRAFV600E has also been linked to worse outcomes and resistance to traditional treatment [9,22].
However, BRAFV600E mutations are comparatively uncommon in breast cancer, although they are more common in some subtypes, such as TNBC [9,12,14]. Despite being less common than in melanoma, BRAFV600E is associated with a more aggressive disease course and has been linked to resistance to treatments such as HER2 inhibitors [10,23]. Given these ramifications, it is more crucial than ever to identify and comprehend BRAFV600E in breast cancer, since it may be a prognostic biomarker and a therapeutic target for more individualised treatment plans [8,12,13,23]. Therefore, further research into the significance of the BRAFV600E mutation in breast cancer is essential to fully understand its effects and improve treatment outcomes for affected individuals [3,9,19].
Methods of Detecting BRAFV600E
There are several methods for detecting the BRAFV600E mutation, which is important in a variety of malignancies. Each method has its own benefits and drawbacks. The three main approaches for detecting BRAFV600E in breast cancer are described in this section: PCR-based methods, Immunohistochemistry (IHC), and sequencing.
Overview of Detection Techniques
Sequencing: DNA sequencing, particularly Next-Generation Sequencing (NGS), offers high accuracy in detecting mutations, including BRAFV600E. It allows for the identification of low-frequency mutations and is suitable for samples with complex genetic backgrounds. However, it can be time-intensive and costly, which limits its routine use in many settings [6].
Immunohistochemistry (IHC): IHC is a widely accessible, cost-effective method that uses BRAFV600E-specific antibodies to detect mutant proteins in tissue samples. However, its sensitivity may vary, particularly in samples with low mutation frequency [24,25].
PCR-based Techniques for BRAFV600E Detection
PCR’s sensitivity, rapidity and affordability make it the method of choice for detecting BRAFV600E mutations. Different PCR techniques have been created and each is appropriate for a particular clinical or scientific context.
Conventional PCR: This is a foundational technique for detecting the BRAFV600E mutation, widely used due to its simplicity and accessibility. It allows for the amplification and detection of target sequences, making it suitable for initial mutation screening. Its advantages include cost-effectiveness, rapid processing and minimal equipment requirements, making it useful in resource-limited settings [3,8,18]. However, it has significant limitations, such as low sensitivity for detecting mutations present at low frequencies, an inability to quantify mutation burden and reduced specificity compared to advanced methods. Consequently, while conventional PCR remains valuable, it is less preferred for precise diagnostic and therapeutic monitoring applications [5,7,18].
Real-Time PCR (qPCR): qPCR enhances conventional PCR by enabling quantitative detection of the BRAFV600E mutation, providing real-time monitoring of DNA amplification. Its key advantages include high sensitivity, which allows for the detection of low-frequency mutations and the ability to quantify mutation levels, making it ideal for clinical diagnostics and monitoring therapeutic response [3,8,11]. Furthermore, qPCR offers faster results and greater accuracy, which are crucial for timely decision-making in treatment strategies. However, it requires more specialised equipment and reagents and its cost is higher than that of conventional PCR, limiting its use in resource-constrained settings [5,7,18].
Droplet digital PCR (ddPCR): This is an advanced method that offers exceptional precision in detecting low-frequency mutations, such as BRAFV600E, by partitioning DNA into thousands of droplets for individual amplification. This technique enhances sensitivity, making it highly effective for detecting rare mutant alleles, even in heterogeneous tumour samples [5,7,18]. ddPCR is particularly valuable for monitoring minimal residual disease, assessing tumour heterogeneity and guiding personalised treatment decisions in breast cancer. Its main advantages include high sensitivity and precision, allowing for accurate mutation quantification. However, ddPCR requires specialised equipment and is more expensive, limiting its widespread use in resource-limited settings [3,11].
Role of BRAFV600E in Breast Cancer
Although it is less frequent in breast cancer, the BRAFV600E mutation is a well-characterised oncogenic change that has been primarily explored in melanoma and papillary thyroid carcinoma [5]. The MAPK/ERK pathway, which supports cellular proliferation and survival, is constitutively activated because of this mutation [5]. Understanding its role in breast cancer is essential due to its potential impact on diagnosis, prognosis, and targeted therapy.
Prevalence of BRAFV600E in breast cancer: Research on BRAFV600E in breast cancer has revealed a range of prevalence rates, indicating that this gene is uncommon in breast cancer in comparison to other malignancies such as melanoma. As an illustration of potential subtype specificity in mutation profiles, certain studies have found low rates or even the absence of the BRAFV600E mutation in breast cancer subtypes, particularly triple-negative and basal-like breast cancers [7,14]. Though BRAFV600E is still uncommon, some research in Asian communities indicates that it might be involved in a subgroup of breast cancers, which calls for more research in a variety of populations [8,17,26].
BRAFV600E as a prognostic marker: Regarding BRAFV600E as a prognostic marker in breast cancer, there is not much agreement. Although the mutation has been linked to worse outcomes in other malignancies, its low incidence in breast cancer makes it unclear what role it plays in prognosis. Larger studies are required to determine the prognostic importance of BRAFV600E within subtypes of breast cancer; however, some case reports and small cohort studies indicate that it may give a more aggressive phenotype [5,21].
BRAFV600E as a therapeutic target in breast cancer: Due to the success of targeting BRAFV600E with certain inhibitors, such as vemurafenib, in melanoma, researchers are now considering its use in breast cancer. Only a small number of case reports show partial responses in individuals with BRAFV600E-mutated metastatic illness, indicating that there is little clinical evidence for BRAF inhibitors in breast cancer [21,23]. Although a combination of BRAF and MEK inhibitors has also demonstrated promise, obstacles still exist because of the low frequency of the mutation and the lack of reliable clinical trials specifically for breast cancer [9,23]. BRAFV600E-targeted treatments may be beneficial for specific patient subgroups or subtypes of breast cancer, depending on additional research.
Clinical Implications of BRAFV600E Status in Breast Cancer
The clinical significance of the BRAFV600E mutation in breast cancer is still being determined, mainly because of its low frequency and inconsistent results in study. But in other situations, knowing its function can be essential to tailoring treatment in other situations. For targeted therapy, BRAFV600E status is important in tumours where it is prevalent, including melanoma, where BRAF inhibitors like vemurafenib are now prominent treatment options [5]. However, the infrequency of the mutation has hindered its integration into standard treatment procedures for breast cancer. Studies have shown that patients with metastatic breast cancer who have the BRAFV600E mutation react to BRAF inhibitors either by themselves or in conjunction with MEK inhibitors, suggesting that this mutation provides a possible avenue for targeted therapy when it occurs [21,23]. These findings imply that, despite their rarity, BRAFV600E-positive breast cancer cases may benefit from treatments that are not typically utilised for this kind of cancer, providing patients with advanced or treatment-resistant disease with additional options.
BRAFV600E’s usefulness as a standard biomarker for breast cancer is limited in several ways. Multiple studies across populations have established its low frequency, which limits the practical use of BRAFV600E-targeted treatments in the management of breast cancer [8,14]. Further complicating its use as a clinical biomarker are conflicting findings about its predictive usefulness in breast cancer.
Although it might occasionally indicate a more aggressive course of the disease, larger studies with data are required to validate these correlations and develop uniform standards for BRAFV600E targeting in breast cancer [5,21]. Given these limitations, more studies are necessary to determine whether a patient’s BRAFV600E status should influence their course of treatment for breast cancer. PCR’s sensitivity and specificity for identifying BRAFV600E in breast cancer are poor, particularly as this mutation is uncommon in this kind of cancer. It is difficult to maintain consistent precision when using high-sensitivity approaches to detect even few BRAFV600E-positive cells in heterogeneous tumours [7]. BRAFV600E inhibitors may work in combination with other targeted treatments, including MEK inhibitors or immunotherapies, to improve outcomes by tackling the mechanisms that lead to tumour growth and resistance [21,23]. Future research should prioritise large studies and clinical trials to determine the prognostic role of BRAFV600E in breast cancer and develop effective targeted therapies. Demographic studies across diverse populations are essential to understand the global impact of this mutation and its relevance in different patient groups.
Conclusion(s)
The low frequency and varying function of BRAFV600E in various studies make it a complicated target for breast cancer, presenting both opportunities and obstacles. PCR is a useful method for identifying this mutation, but its wider use in breast cancer diagnosis is constrained by problems with sensitivity and specificity. Research indicates that BRAFV600E may be a therapeutic target and have prognostic significance when paired with other inhibitors. Extensive research is needed to determine the clinical significance of this rare mutation in breast cancer across diverse populations. Future studies should concentrate on developing conclusive treatment procedures, improving detection techniques and investigating combination therapies to fully realise the promise of BRAFV600E in improved patient outcomes.
[1]. Malvia S, Bagadi SA, Dubey US, Saxena S, Epidemiology of breast cancer in Indian womenAsia Pac J Clin Oncol 2017 13(4):289-95.Epub 2017 Feb 910.1111/ajco.1266128181405 [Google Scholar] [CrossRef] [PubMed]
[2]. Mehrotra R, Yadav K, Breast cancer in India: Present scenario and the challenges aheadWorld J Clin Oncol 2022 13(3):209-18.10.5306/wjco.v13.i3.20935433294PMC8966510 [Google Scholar] [CrossRef] [PubMed]
[3]. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Mutations of the BRAF gene in human cancerNature 2002 417(6892):949-54.Epub 2002 Jun 910.1038/nature0076612068308 [Google Scholar] [CrossRef] [PubMed]
[4]. Geyer FC, Marchio C, Reis-Filho JS, The role of molecular analysis in breast cancerPathology 2009 41(1):77-88.10.1080/0031302080256353619089743 [Google Scholar] [CrossRef] [PubMed]
[5]. Cantwell-Dorris ER, O’Leary JJ, Sheils OM, BRAFV600E: Implications for carcinogenesis and molecular therapyMol Cancer Ther 2011 10(3):385-94.10.1158/1535-7163.MCT-10-079921388974 [Google Scholar] [CrossRef] [PubMed]
[6]. Cancer Genome Atlas NetworkComprehensive molecular portraits of human breast tumoursNature 2012 490(7418):61-70.Epub 2012 Sep 2310.1038/nature1141223000897PMC3465532 [Google Scholar] [CrossRef] [PubMed]
[7]. Myers MB, Banda M, McKim KL, Wang Y, Powell MJ, Parsons BL, Breast cancer heterogeneity examined by high-sensitivity quantification of PIK3CA, KRAS, HRAS, and BRAF mutations in normal breast and ductal carcinomasNeoplasia 2016 18(4):253-63.10.1016/j.neo.2016.03.00227108388PMC4840288 [Google Scholar] [CrossRef] [PubMed]
[8]. Wang YL, Dai X, Li YD, Cheng RX, Deng B, Geng XX, Study of PIK3CA, BRAF, and KRAS mutations in breast carcinomas among Chinese women in QinghaiGenet Mol Res 2015 14(4):14840-46.10.4238/2015.November.18.4926600545 [Google Scholar] [CrossRef] [PubMed]
[9]. Rocca A, Braga L, Volpe MC, Maiocchi S, Generali D, The predictive and prognostic role of RAS-RAF-MEK-ERK pathway alterations in breast cancer: Revision of the literature and comparison with the analysis of cancer genomic datasetsCancers (Basel) 2022 14(21):530610.3390/cancers1421530636358725PMC9653766 [Google Scholar] [CrossRef] [PubMed]
[10]. Patra S, Young V, Llewellyn L, Senapati JN, Mathew J, BRAF, KRAS and PIK3CA mutation and sensitivity to Trastuzumab in breast cancer cell line modelAsian Pac J Cancer Prev 2017 18(8):2209-13.10.22034/APJCP.2017.18.8.220928843257PMC5697482 [Google Scholar] [CrossRef] [PubMed]
[11]. Ueda M, Toji E, Nunobiki O, Izuma S, Okamoto Y, Torii K, Mutational analysis of the BRAF gene in human tumour cellsHum Cell 2008 21(2):13-17.10.1111/j.1749-0774.2008.00046.x18397470 [Google Scholar] [CrossRef] [PubMed]
[12]. Mahesh K, Ch KK, Ravi Kanth K, Laxmi Addala VV, Murthy S, Differences in gene expression profiles between human breast tissue and peripheral blood samples for breast cancer detectionJ Cancer Sci Ther 2012 4:379-85. [Google Scholar]
[13]. Grob TJ, Heilenkötter U, Geist S, Paluchowski P, Wilke C, Jaenicke F, Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancerBreast Cancer Res Treat 2012 134(2):561-67.10.1007/s10549-012-2092-722610646 [Google Scholar] [CrossRef] [PubMed]
[14]. Tilch E, Seidens T, Cocciardi S, Reid LE, Byrne D, Simpson PT, Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian womenBreast Cancer Res Treat 2014 143(2):385-92.Epub 2013 Dec 710.1007/s10549-013-2798-124318467 [Google Scholar] [CrossRef] [PubMed]
[15]. Kim WG, Jung HK, Kim YM, Kim WW, Lee JS, Kim SJ, BRAF mutation from tissue samples in Korean patients with breast cancer and thyroid cancer: A pilot studyJ Korean Society for Breast Screening 2015 12(1):28-34. [Google Scholar]
[16]. Wang L, Lu Q, Jiang K, Hong R, Wang S, Xu F, BRAF V600E mutation in triple-negative breast cancer: A case report and literature reviewOncol Res Treat 2022 45(1-2):54-61.Epub 2021 Nov 2410.1159/00052045334818649PMC8985016 [Google Scholar] [CrossRef] [PubMed]
[17]. Al-Askeri MA, Mutter AA, Mutation of BRAF V600E in Iraqi female patients diagnosed with breast cancerJournal of University of Babylon for Pure and Applied Sciences 2018 26(3):78-83. [Google Scholar]
[18]. Eachkoti R, Farooq S, Reshi R, Rehman MU, Rashid T, Naikoo NA, Mutation allele-specific multiplex PCR for the detection of BRAFV600E mutations in breast carcinomasWorld Academy Sci J 2019 1(3):145-50.10.3892/wasj.2019.14 [Google Scholar] [CrossRef]
[19]. Sholl LM, A narrative review of BRAF alterations in human tumours: Diagnostic and predictive implicationsPrecis Cancer Med 2020 3:2610.21037/pcm-20-39 [Google Scholar] [CrossRef]
[20]. Rashid FA, Munkhdelger J, Fukuoka J, Bychkov A, Prevalence of BRAFV600E mutation in Asian series of papillary thyroid carcinoma-A contemporary systematic reviewGland Surg 2020 9(5):1878-900.10.21037/gs-20-43033224863PMC7667088 [Google Scholar] [CrossRef] [PubMed]
[21]. Pircher M, Winder T, Trojan A, Response to Vemurafenib in metastatic triple-negative breast cancer harbouring a BRAF V600E mutation: A case report and electronically captured patient-reported outcomeCase Rep Oncol 2021 14(1):616-21.10.1159/00051390533976643PMC8077504 [Google Scholar] [CrossRef] [PubMed]
[22]. Abuali I, Lee CS, Seetharamu N, A narrative review of the management of BRAF non-V600E mutated metastatic non-small cell lung cancerPrecis Cancer Med 2022 5:1310.21037/pcm-21-49 [Google Scholar] [CrossRef]
[23]. López de Sá A, de Luna A, Antoñanzas M, García-Barberán V, Moreno-Anton F, García-Sáenz JA, Case report: Clinical success targeting BRAF-mutated, hormone receptor positive, HER2- negative advanced breast cancer patient with BRAF-inhibitor plus MEK- inhibitorFront Oncol 2022 12:99734610.3389/fonc.2022.99734636531075PMC9755882 [Google Scholar] [CrossRef] [PubMed]
[24]. Khan S, Bhake A, Sagar S, Deciphering the role of BRAFV600E immunohistochemistry in breast lesions: A comprehensive reviewCureus 2024 16(7):e6487210.7759/cureus.6487239156294PMC11330685 [Google Scholar] [CrossRef] [PubMed]
[25]. Cui G, Liu D, Li W, Fu X, Liang Y, Li Y, A meta-analysis of the association between BRAF mutation and non small cell lung cancerMedicine (Baltimore) 2017 96(14):e655210.1097/MD.000000000000655228383426PMC5411210 [Google Scholar] [CrossRef] [PubMed]
[26]. Jung YY, Jung WH, Koo JS, BRAF mutation in breast cancer by BRAF V600E mutation-specific antibodyInt J Clin Experi Pathol 2016 9(2):1545-56. [Google Scholar]