Introduction
Matrix Metalloproteinase 3 (MMP3) is a vital member of the MMP family, known for its wide range of substrate specificity and proteolytic activity against Extracellular Matrix (ECM) components. The role of functional polymorphisms in the MMP genes has been previously investigated in relation to cancer susceptibility, particularly breast cancer. Several Single-Nucleotide Polymorphisms (SNPs) in the MMP3 gene have been linked to a number of clinical illnesses, such as Coronary Artery Disease (CAD); however, the results were not entirely conclusive.
Aim
To identify pathogenic missense SNPs in the human MMP3 gene and analyse their effects on structure and function.
Materials and Methods
This was a record-based cross-sectional study performed using data retrieved from online resources. The analysis was conducted using a series of different bioinformatic tools, for which ethical clearance was obtained from the institution. The online tools used included Sorting Intolerant from Tolerant (SIFT), PolyPhen-2, PhD-SNP, PANTHER, PROVEAN, and SNPs and GO to predict harmful non synonymous SNPs (nsSNPs). Further analysis was performed using I-Mutant 2.0, MutPred2, Consurf, and HOPE software. These tools were able to filter out damaging SNPs and predict the impact of deleterious SNPs on the structure and function of the MMP3 protein.
Results
This study predicted two potentially pathogenic SNPs (D175Y and Y116C) out of 443 missense SNPs from dbSNP, which is a database of SNPs available on the National Centre for Biotechnology Information website. Further analysis revealed that these SNPs were located in highly conserved regions and were predicted to decrease protein stability.
Conclusion
In this study, two potentially pathogenic SNPs (D175Y and Y116C) were identified. Characterisation of these SNPs can help us gain a better understanding of the molecular basis of clinical conditions. The results of this study can be further validated by designing population-based studies and wet lab experiments. This will help in augmenting research and personalised medicine.
Introduction
The human genome is relatively large, comprising approximately six billion nucleotides. About 99.9% of the DNA sequences in the human genome are the same across individuals, with individual variations accounting for the remaining 0.1% [1]. Single Nucleotide Polymorphisms (SNPs), which refer to the substitution of a single nucleotide, are the most prevalent type of genetic variation. SNPs occur throughout the genome at a rate of one in every 1,000 base pairs. SNPs are broadly categorised into two types: synonymous and non synonymous (nsSNP). Non synonymous SNPs are responsible for protein variations and amino acid substitutions in humans, while synonymous SNPs do not change the amino acid sequences [2]. With the development of practical and readily accessible DNA sequencing tools, SNPs have gained substantial clinical significance over the past two decades. Regardless of their effects on the biological function of gene products, SNPs are increasingly recognised as valuable markers in mapping complex human diseases, population genetics, and evolutionary studies [3]. Efficient identification of important SNPs and understanding their impact on protein function will be beneficial for genetic studies aimed at elucidating the molecular basis of various diseases. Furthermore, targeted in-vitro and in-vivo experiments can be designed to predict the effects of specific SNPs [4]. In light of this, multiple studies are being conducted to predict the functional consequences of nsSNPs, particularly based on sequence information and structural attributes [5-10].
Matrix Metalloproteinases (MMPs) are a family of more than 25 zinc-containing enzymes that break down various proteoglycans, fibronectin, interstitial collagen, and basement membrane collagen during normal physiological remodelling and repair processes in development and inflammation [11]. MMP-3, also known as Stromelysin-1, is a crucial member of the MMP family that exhibits a wide range of substrate specificity. Additionally, MMP-3 plays a critical role in the remodelling of connective tissue by activating other MMPs, such as collagenase, matrilysin, and gelatinase B [12,13]. The role of MMP gene functional polymorphisms in cancer susceptibility, especially breast cancer, has been previously investigated. The 5A/6A polymorphism has been linked to patient survival, oestrogen receptor status, breast cancer risk, and tumour size based on multiple genome-wide association analyses [14]. However, the association between SNPs and breast cancer risk remains inconclusive and controversial, as it is largely influenced by the ethnicity of the population and sample size [14-16]. Apart from breast cancer, a meta-analysis indicates a significant correlation between gastrointestinal, colorectal, and oesophageal malignancies and the MMP-3-1171 5A/6A variation [17].
In addition to malignancies, MMP3 gene polymorphisms have been found to be associated with atherosclerosis. This is due to the critical role of MMP3 in the degradation of the Extracellular Matrix (ECM) of blood vessels. Several studies evaluating the influence of promoter polymorphism and MMP-3 (-1612 5A/6A) on CAD risk have produced contradictory results, including gender differences as well [18-20]. Two MMP3 polymorphisms, rs520540 A/G and rs679620 C/T, were associated with cerebral stroke risk; however, contrary results were obtained in a different population, probably due to racial differences [21].
Among these reported SNPs, distinguishing functional SNPs from neutral SNPs is a major challenge. Experimental wet lab studies aimed at identifying the effects of multiple SNPs are time-consuming, challenging, and costly. Applying computational approaches to filter out possibly detrimental SNPs is emerging as a novel strategy for understanding the molecular mechanisms of diseases [22].
This was an in-silico study utilising bioinformatics computational tools with good predictive capabilities to identify the possible deleterious SNPs in the MMP-3 gene. Several studies have been conducted to find the association between specific SNPs in the MMP3 gene and diseases [14-21]. However, according to the literature search, there have been no comparable in-silico studies that examined every SNP in MMP-3 from the NCBI database. This study is expected to unravel the potential impact of polymorphisms on MMP3 structure and function. In addition, it can help in designing further specific experiments to confirm the importance of these SNPs in various clinical conditions. Combined with experimental data, it can aid in the development of personalised medicine.
Materials and Methods
This online records-based cross-sectional study was conducted at Government Medical College, Manjeri, Kerala, India over a period from May to July 2024 (three months), which included data collection as well as analysis using online computational tools. IEC approval for this study was obtained from the institution (Ref. No: IEC/GMCM/133, date: 18/05/2024).
Methodology
The operational flow chart of this study is presented in [Table/Fig-1]. The detailed methodology is as follows:
Operational flow chart for in-silico analysis of missense SNPs in MMP3 gene.
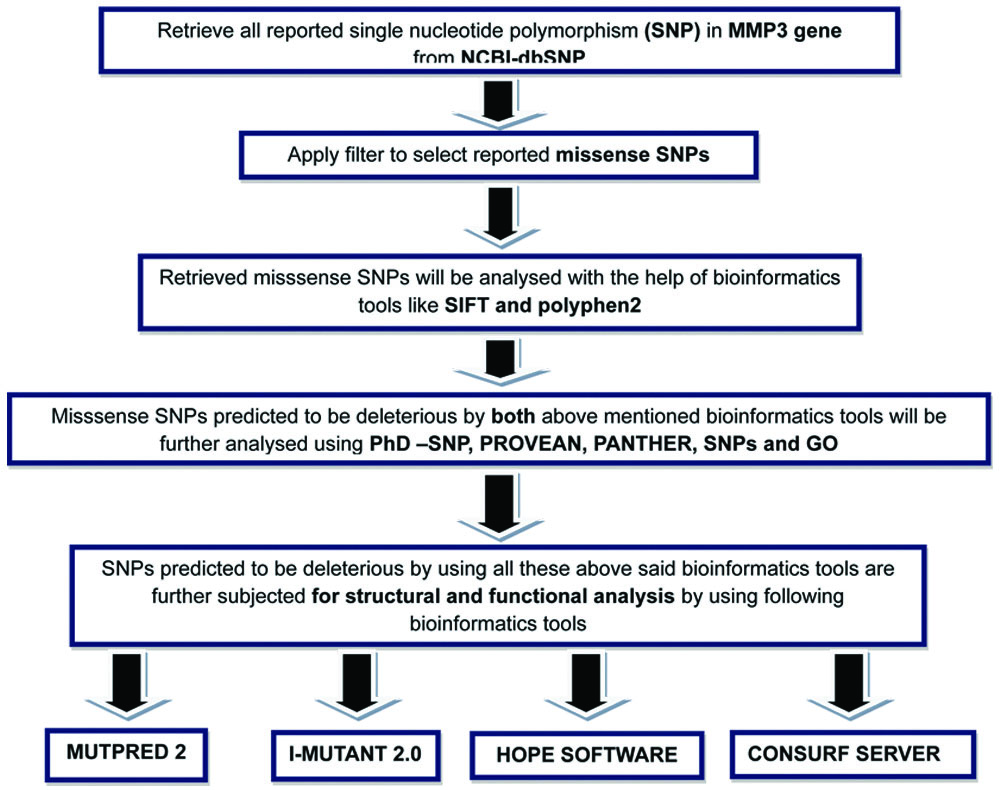
Retrieval of SNPs in the MMP3 gene: The ncbi SNPs database (dbSNP) (https://www.ncbi.nlm.nih.gov/snp/) was used to retrieve all known SNPs for MMP-3 in humans [23]. Non synonymous (missense) mutations were selected from the dataset for further analysis. The UniProt database was utilised to obtain the protein’s sequence and other pertinent information.
Prediction of the most deleterious missense SNPs: The retrieved missense SNPs were analysed using bioinformatics-based web tools for identification of high-risk nsSNPs. Details of the tools used are provided in [Table/Fig-2]. Polymorphism phenotyping v2 (PolyPhen-2) and SIFT were the initial web-based tools employed. A SIFT score value between 0 and 1 indicates a potentially harmful effect, with scores below 0.05 being particularly concerning [24]. A PolyPhen score ranging from 0.85 to 1 is considered probably damaging [25,26]. SNPs identified as potentially harmful by the aforementioned tools were further subjected to additional bioinformatics tools for confirmation and further refinement [27]. The tools used included Predictions from Predictor of Human Deleterious Single Nucleotide Polymorphisms (PhD-SNP) [28], PANTHER-position-specific evolutionary preservation [29], PROVEAN [30], and SNPs and GO [31]. SNPs projected by every tool to be harmful were selected for further examination.
Details of the study’s bioinformatics tools.
| S. No. | Tool | Website | Based on | Input | Output |
|---|
| 1 | SIFT | https://sift.bii.a-star.edu.sg/ | Sequence homology and the physical properties of amino acids | rsID | Score and prediction |
| 2 | Polyphen2 | http://genetics.bwh.harvard.edu/pph2/ | Structural and comparative evolutionary considerations | unitprotID or fasta protein sequence, amino acid change | Score and prediction |
| 3 | Phd-SNP | http://snps.biofold.org/phd-snp/phd-snp.html | Supervised learning based | FASTA sequence, swiss-prot code, amino acid change | RI value and prediction |
| 4 | PANTHER | https://www.pantherdb.org/ | Sequence homology | FASTA sequence and substitutions | PSEP score and message |
| 5 | PROVEAN | http://provean.jcvi.org/index.php | Sequence homology | rsID | Score and prediction |
| 6 | SNPs&GO | https://snps.biofold.org/snps-and-go/snps-and-go.html | Supervised learning based | FASTA sequence, swiss-prot code, amino acid change | RI value and prediction |
| 7 | MUTPRED2 | http://mutpred.mutdb.org | Machine learning-based method | Fasta sequence and amino acid change | Pathogenicity prediction, and a ranked list of specific molecular alterations |
| 8 | I-MUTANT | https://folding.biofold.org/i-mutant/ | Support vector machine -based web server | Protein sequence and amino acid change | DDG value and reliability index |
| 9 | CONSURF | https://consurf.tau.ac.il | Reconstruction of phylogenetic tree reflecting evolutionary relations | Uniprot ID | Amino acid sequence conservation scores |
| 10 | HOPE | https://www3.cmbi.umcn.nl/hope | BLAST, Homology modelling, information from Uniprot and Reprof | Unitprot ID, Fasta sequence and amino acid change | Effect of SNP on protein structure |
| 11. | MUTATION3D | http://www.mutation3d.org/ | Complete-linkage clustering | unitprotID, amino acid change | 3D structure and linear protein models |
Structural and functional analysis of SNPs with predicted damaging consequences:Identification of impact on the structural and functional properties of MMP3: MutPred2 is a web application that integrates genetic and molecular data. It provides a ranked list of specific molecular changes with potential impacts on phenotype, along with a general pathogenicity prediction [32].
Prediction of change in stability of the MMP3 protein using I-Mutant 2.0 Suite: The influence of nsSNPs on protein stability was assessed using the I-Mutant 2.0, which utilises the free energy change value (Delta Delta G: DDG). The DDG represents the difference between the free energy of the folded wild-type and mutant structures. DDG values fall into three categories: neutral (−0.5≤DDG≤0.5 kcal/mol), mostly stable (DDG >0.5 kcal/mol), or highly unstable (DDG <−0.5 kcal/mol) [33].
Prediction of evolutionary conservation of MMP3 using the ConSurf server: The ConSurf web server was used to examine the evolutionary rate of conservation of amino acids in MMP3. ConSurf determines the conservation scores of the protein’s amino acid positions by analysing the evolutionary relationships among similar sequences. The highest conserved location is graded as 9, while conservation values range from 1 to 9 [34].
Protein structure prediction and mutant analysis using HOPE software: The web-based application Have (y) Our Protein Explained (HOPE) is used to analyse how SNPs affect the structural conformation and functionality of proteins. HOPE uses data from multiple web services and databases to generate reports regarding the impact of SNPs, which include text, graphs, and animations [35].
Results
Data on SNPs retrieved: In the MMP3 gene, the dbSNP yielded a total of 4,120 SNPs. Among these SNPs, 443 were missense, 196 were synonymous, and 2,720 were located in the intronic region. For further in-silico research, only the missense or nsSNPs were selected. The remaining 761 SNPs belonged to other functional classes, for which authors were unable to retrieve information from dbSNP. The functional classes in dbSNP delineate the position of a polymorphism in relation to identifiable features of a specific gene transcript. Most functional classes are determined by the location of the variation in relation to the exon boundaries of the transcript [23]. Since present study requires only missense SNPs, only those were retrieved and analysed. The remaining 3,677 SNPs were excluded from the manuscript due to our inability to obtain accurate information about their functional class.
Identification of predicted deleterious SNPs: The selected missense SNPs were subjected to SIFT and PolyPhen-2 to identify possible deleterious SNPs. Out of the 443 SNPs, 14 were predicted to be damaging by both SIFT and PolyPhen-2 [Table/Fig-3] [24-26]. All 14 shortlisted SNPs identified by these two web-based techniques were further analysed using other bioinformatics tools: PhD-SNP, PANTHER, PROVEAN, and SNPs and GO [Table/Fig-4] [28-31]. Among all the analysed SNPs, two nsSNPs, rs201427128 (D175Y) and rs373783506 (Y116C), were predicted to be damaging by all six bioinformatics tools, including SIFT and PolyPhen.
Results of SIFT and Polyphen2 bioinformatics tools.
| S. No. | SNP rsID | Amino acid change | SIFT score | Polyphen-2 score |
|---|
| 1 | rs185081958 | A391V | 0.012 | 0.999 |
| 2 | rs373335227 | Y401H | 0.001 | 0.992 |
| 3 | rs201409153 | Y344C | 0.008 | 0.959 |
| 4 | rs375381761 | L294W | 0 | 0.997 |
| 5 | rs147533686 | R316C | 0 | 1 |
| 6 | rs145941285 | I159T | 0.001 | 0.976 |
| 7 | rs150552863 | I220T | 0.004 | 0.938 |
| 8 | rs201427128 | D175Y | 0.015 | 0.92 |
| 9 | rs140794544 | P187S | 0 | 0.993 |
| 10 | rs141650167 | R91W | 0 | 1 |
| 11 | rs61753776 | W109G | 0 | 0.996 |
| 12 | rs142984123 | Y116C | 0.01 | 1 |
| 13 | rs373783506 | V144M | 0.001 | 0.994 |
| 14 | rs374032529 | L10P | 0.003 | 0.919 |
Results of SNPs chosen to be damaging by SIFT and Polyphen when subjected to other bioinformatics tools.
| Amino acid change | Phd-SNP | PROVEAN | PANTHER | SNPs and GO |
|---|
| Prediction | RI | Score | Prediction | Preservation time | Message | Pdel | Prediction | RI |
|---|
| A391V | Neutral | 7 | -3.661 | Deleterious | 911 | Probably damaging | 0.85 | Neutral | 6 |
| Y401H | Disease | 4 | -4.248 | Deleterious | 361 | Possibly damaging | 0.5 | Neutral | 6 |
| Y344C | Disease | 5 | -7.689 | Deleterious | 455 | Probably damaging | 0.57 | Neutral | 6 |
| L294W | Neutral | 7 | -5.189 | Deleterious | 456 | Probably damaging | 0.57 | Neutral | 5 |
| R316C | Neutral | 1 | -7.293 | Deleterious | 797 | Probably damaging | 0.74 | Neutral | 1 |
| I159T | Disease | 1 | -4.934 | Deleterious | 911 | Probably damaging | 0.85 | Neutral | 4 |
| I220T | Disease | 5 | -2.424 | Neutral | 176 | Probably benign | 0.27 | Neutral | 8 |
| D175Y | Disease | 5 | -8.686 | Deleterious | 911 | Probably damaging | 0.85 | Disease | 3 |
| P187S | Disease | 2 | -7.858 | Deleterious | 911 | Probably damaging | 0.85 | Neutral | 2 |
| R91W | Neutral | 1 | -7.527 | Deleterious | 911 | Probably damaging | 0.85 | Disease | 3 |
| W109G | Neutral | 1 | -12.841 | Deleterious | 911 | Probably damaging | 0.85 | Disease | 3 |
| Y116C | Disease | 3 | -8.879 | Deleterious | 911 | Probably damaging | 0.85 | Disease | 2 |
| V144M | Disease | 2 | -2.593 | Deleterious | 797 | Probably damaging | 0.74 | Neutral | 5 |
| L10P | Disease | 3 | -4.604 | Deleterious | 176 | Probably benign | 0.27 | Neutral | 3 |
Prediction by MutPred2 on structural and functional effects [32]: The selected two detrimental SNPs were analysed using MutPred2, which suggested a significant impact on the 3D structure of the MMP3 protein [Table/Fig-5].
Results from MutPred analysis.
| SNP | Molecular changes (p-values <=0.05) | MutPred score |
|---|
| D175Y | Altered metal binding, ordered interface and Transmembrane proteinLoss of Catalytic site at D175 Loss of Relative solvent accessibility Loss of Allosteric site at F174 Loss of Sulfation at Y172 | 0.921 |
| Y116C | Alteration in ordered interface, Metal binding, DNA binding, Transmembrane protein Loss of Allosteric site at Y121 Altered stability | 0.849 |
Prediction of changes in protein stability by I-Mutant 2.0 [33]: I-Mutant predicted the extent to which the damaging SNPs alter the stability of the MMP3 protein. Both SNPs were predicted to decrease the stability of the protein [Table/Fig-6].
Results of I-mutant 2.0 analysis of nsSNPs.
| S. No. | SNP | DDG (Kcal/mol) | RI |
|---|
| 1 | rs201427128 (D175Y) | -1.40 | 3 |
| 2 | rs373783506 (Y116C) | -0.03 | 2 |
Consurf server results [34]: An analysis of conservation using the ConSurf server revealed that both SNPs, D175Y and Y116C, were located in highly conserved areas and were found to be exposed residues [Table/Fig-7].
ConSurf results for MMP3 protein.

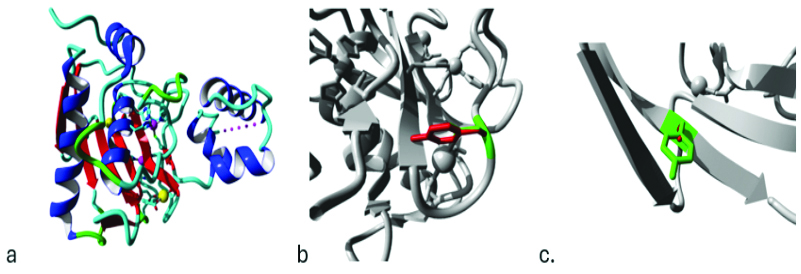
Predicted changes in the 3D structure of the MMP3 protein due to nsSNPs [35]: HOPE software was used to generate results for both nsSNPs, with structural information obtained from the Protein Data Bank (PDB) database. The PDB ID used was 1SLM. In the case of D175Y, the mutant residue is larger, impacting hydrogen bond formation and its interaction with the metal ion in the original wild-type residue. Additionally, there is a disturbance in the ionic interaction made by the wild-type residue. For Y116C, the smaller residue results in an empty space in the protein core, which disrupts proper folding by causing a loss of hydrogen bonds in the core [Table/Fig-8].
a) An overview of the protein is presented in a ribbon format, with colours indicating different structural elements: α-helix in blue, β-strand in red, turn in green, 3/10 helix in yellow, and random coil in cyan; b) A close-up view of the D175Y mutation; c) A close-up view of the Y116C mutation. The protein is rendered in grey, while the side chains of both the wild-type and mutant residues are highlighted in green and red, respectively.
Discussion
The MMP-3 gene, which codes for the MMP-3 protein, is a component of the MMP gene cluster located on chromosome 11q22.3. The NCBI gene ID for MMP3 is 4314, and its UniProt ID is P08254 [36,37]. The expression of the MMP3 gene may be regulated by DNA polymorphisms, which can exert allele-specific effects, resulting in fundamental variations in each person’s susceptibility to diseases and other traits [11].
The goal of the current study was to use several computational techniques to discover SNPs that could be harmful and alter the structure or function of the MMP3 gene. A significant number of researchers have utilised in-silico tools to predict the structural and functional impact of SNPs on proteins, such as the human CTLA4 gene [38], IL-33 gene [39], and the androgen receptor gene [27]. This work represents the first comprehensive and systematic in-silico study of functional SNPs in the MMP3 gene.
Combining multiple computational methods can enhance the validity of predictions regarding damaging SNPs and improve efficiency. By employing six prediction methods (SIFT, PolyPhen-2, PROVEAN, PANTHER, PhD-SNP, and SNPs and GO), authors were able to obtain an integrated view of harmful SNPs in the MMP3 gene. Since each method relies on a different set of parameters, two ns SNPs (D175Y and Y116C) were identified as high-risk in present study, which were predicted to be harmful by every algorithm. Additional research revealed that these two ns SNPs might be dangerous because they are located in a highly conserved region and have the potential to affect protein stability [6-9,22,27,38,39].
As an important member of the MMP3 family, several experimental studies have been conducted on genetic variation in MMP3, especially in malignancies [11,13-18,21]. Wang L et al., conducted an observational study and meta-analysis of MMP3 gene polymorphism in the Chinese population. The researchers concluded that while MMP-3 gene polymorphism raises the risk of ovarian cancer in the southern Chinese population, their meta-analysis showed that it had no effect on risk in other populations [40]. Durmanova V et al., demonstrated that the MMP3 SNP rs3025058 influences the age of onset of Alzheimer’s disease [41].
The predicted ns SNPs from present study have not been linked to any clinical disease in published studies. Therefore, to support this finding, it is necessary to validate these ns SNPs through larger population-based studies and specifically designed wet lab experiments.
Limitation(s)
The present study utilised the limited dataset from the NCBI dbSNP database. More ns SNPs could have been analysed if other databases had been included.
Conclusion(s)
Using a variety of bioinformatics techniques, this study has identified two significant nsSNPs, D175Y and Y116C, from the extensive SNP dataset of MMP3. Based on these findings, it can be concluded that these SNPs should be regarded as significant candidates for planning functional experiments involving the MMP3 gene. This presents an opportunity to investigate the molecular basis of any associated clinical problems and may aid in the discovery of potential drugs or pharmacological targets. Although computational biology is powerful, it has its limitations; therefore, population-based research is necessary to provide definitive validation.
[1]. Teama S, DNA Polymorphisms: DNA-Based molecular markers and their application in medicine [Internet]Genetic Diversity and Disease Susceptibility 2018 InTechAvailable from: https://dx.doi.org/10.5772/intechopen.7951710.5772/intechopen.79517 [Google Scholar] [CrossRef]
[2]. Hossain MS, Roy AS, Islam MS, In silico analysis predicting effects of deleterious SNPs of human RASSF5 gene on its structure and functionsSci Rep 2020 10(1):1454210.1038/s41598-020-71457-132884013 [Google Scholar] [CrossRef] [PubMed]
[3]. Wakui M, Analysis of Single Nucleotide Polymorphisms (SNPs)Rinsho Byori 2013 61(11):1008-17. [Google Scholar]
[4]. Chitrala KN, Yeguvapalli S, Computational screening and molecular dynamic simulation of breast cancer associated deleterious non-synonymous single nucleotide polymorphisms in TP53 genePloS one 2014 9(8):e10424210.1371/journal.pone.010424225105660 [Google Scholar] [CrossRef] [PubMed]
[5]. George Priya Doss C, Rajasekaran R, Sudandiradoss C, Ramanathan K, Purohit R, Sethumadhavan R, A novel computational and structural analysis of nsSNPs in CFTR geneGenomic Med 2008 2(1-2):23-32.10.1007/s11568-008-9019-818716917 [Google Scholar] [CrossRef]
[6]. Nouira S, Bouatrous E, Ouragini H, An in-silico analysis of the coding and non-coding non-synonymous single nucleotide variants in the Npm1 gene to determine susceptibility to cancerSSRN 2024 Available from: https://doi.org/10.2139/ssrn.487155010.2139/ssrn.4871550 [Google Scholar] [CrossRef]
[7]. Almakhari M, Chen Y, Kong AS, Moradigaravand D, Lai KS, Lim SH, In-silico identification of deleterious non-synonymous SNPs of TBX1 gene: Functional and structural impact towards 22q11. 2DSPlos one 2024 19(6):e029809210.1371/journal.pone.029809238905172 [Google Scholar] [CrossRef] [PubMed]
[8]. Subramanian S, Sanjeev K, Prakash PS, Appukuttan D, In silico analysis of single nucleotide polymorphisms of NAT-2 gene and its implications in periodontitisInt J Chem Biochem Sci 2023 24(8):177-81. [Google Scholar]
[9]. Wiguna NY, Hawa P, Jonny J, Human PPARG gene mutation as risk factor for Type 2 diabetes melitus: In silico analysisASEAN Journal of Military and Preventive Medicine 2024 1(1):01-02. [Google Scholar]
[10]. Abdelazim AA, Maged M, Abdelmaksoud AI, Hassanin SE, In-silico analysis of nsSNPs in human CYP3A4/5 and their effects on drug-enzymes interactions of FDA-approved COVID-19 antiviral drugs: A pharmacogenetic studyResearch Square 2024 10.21203/rs.3.rs-3910067/v1 [Google Scholar] [CrossRef]
[11]. Munhoz Godoy-Santos AL, Santos MC, MMP-3 polymorphism: Genetic marker in pathological processesMol Med Rep 2010 3(5):735-40. [Google Scholar]
[12]. Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA, Guerrero-Rodriguez JF, The roles of matrix metalloproteinases and their inhibitors in human diseasesInt J Mol Sci 2020 21(24):973910.3390/ijms2124973933419373 [Google Scholar] [CrossRef] [PubMed]
[13]. Ye S, Eriksson P, Hamsten A, Kurkinen M, Humphries SE, Henney AM, Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expressionJ Biol Chem 1996 271(22):13055-60.10.1074/jbc.271.22.130558662692 [Google Scholar] [CrossRef] [PubMed]
[14]. Suhaimi SA, Chan SC, Rosli R, Matrix Metallopeptidase 3 polymorphisms: Emerging genetic markers in human breast cancer metastasisJ Breast Cancer 2020 23(1):01-09.10.4048/jbc.2020.23.e1732140265 [Google Scholar] [CrossRef] [PubMed]
[15]. Peng B, Cao L, Wang W, Xian L, Jiang D, Zhao J, Polymorphisms in the promoter regions of matrix metalloproteinases 1 and 3 and cancer risk: A meta-analysis of 50 case-control studiesMutagenesis 2010 25(1):41-48.10.1093/mutage/gep04119843588 [Google Scholar] [CrossRef] [PubMed]
[16]. Ghilardi G, Biondi ML, Caputo M, Leviti S, DeMonti M, Guagnellini E, A single nucleotide polymorphism in the matrix metalloproteinase-3 promoter enhances breast cancer susceptibilityClin Cancer Res 2002 8(12):3820-23. [Google Scholar]
[17]. Aziz MA, Jafrin S, Barek MA, Anonna SN, Islam MS, MMP-3 -1171 5A/6Apromoter polymorphism and cancer susceptibility: An updated meta-analysis and trial sequential analysisFuture Oncol 2023 19(21):1495-512.10.2217/fon-2022-130637551683 [Google Scholar] [CrossRef] [PubMed]
[18]. Wenwang L, Susceptibility of MMP3 gene polymorphism to coronary artery disease: A meta-analysisJ Med Biochem 2023 42(4):685-93.10.5937/jomb0-4331538084252 [Google Scholar] [CrossRef] [PubMed]
[19]. Beyzade S, Zhang S, Wong YK, Day IN, Eriksson P, Ye S, Influences of matrix metalloproteinase-3 gene variation on extent of coronary atherosclerosis and risk of myocardial infarctionJ Am Coll Cardiol 2003 41(12):2130-37.10.1016/S0735-1097(03)00482-012821236 [Google Scholar] [CrossRef] [PubMed]
[20]. Koch W, de Waha A, Hoppmann P, Schömig A, Kastrati A, Haplotypes and 5A/6A polymorphism of the matrix metalloproteinase-3 gene in coronary disease: Case-control study and a meta-analysisAtherosclerosis 2010 208(1):171-76.10.1016/j.atherosclerosis.2009.08.02119762026 [Google Scholar] [CrossRef] [PubMed]
[21]. Yin Y, Zhang Y, Zhang X, Zhang Q, Wang J, Yang T, Association of MMP3, MMP14, and MMP25 gene polymorphisms with cerebral stroke risk: A case-control studyBMC Med Genomics 2023 16(1):29710.1186/s12920-023-01734-137986083 [Google Scholar] [CrossRef] [PubMed]
[22]. Desai M, Chauhan J, In silico analysis of nsSNPs in human methyl CpG binding protein 2Meta Gene 2016 10:01-07.10.1016/j.mgene.2016.09.004 [Google Scholar] [CrossRef]
[23]. Bhagwat M, Searching NCBI’s dbSNP databaseCurr Protoc Bioinformatics 2010 32(1):01-09.10.1002/0471250953.bi0119s3221154707 [Google Scholar] [CrossRef] [PubMed]
[24]. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC, SIFT web server: Predicting effects of amino acid substitutions on proteinsNucleic Acids Res 2012 40(W1):W452-57.10.1093/nar/gks53922689647 [Google Scholar] [CrossRef] [PubMed]
[25]. Adzhubei I, Jordan DM, Sunyaev SR, Predicting functional effect of human missense mutations using PolyPhen-2Curr Protoc Hum Genet 2013 76(1):07-20.10.1002/0471142905.hg0720s7623315928 [Google Scholar] [CrossRef] [PubMed]
[26]. Gemovic B, Perovic V, Glisic S, Veljkovic N, Feature-based classification of amino acid substitutions outside conserved functional protein domainsScientific World J 2013 2013(1):94861710.1155/2013/94861724348198 [Google Scholar] [CrossRef] [PubMed]
[27]. Adiba M, Das T, Paul A, Das A, Chakraborty S, Hosen MI, In silico characterization of coding and non-coding SNPs of the androgen receptor geneInformatics in Medicine Unlocked 2021 24:10055610.1016/j.imu.2021.100556 [Google Scholar] [CrossRef]
[28]. Capriotti E, Calabrese R, Casadio R, Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary informationBioinformatics 2006 22(22):2729-34.10.1093/bioinformatics/btl42316895930 [Google Scholar] [CrossRef] [PubMed]
[29]. Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, Mi H, PANTHER: Making genome-scale phylogenetics accessible to allProtein Sci 2022 31(1):08-22.10.1002/pro.421834717010 [Google Scholar] [CrossRef] [PubMed]
[30]. Choi Y, Chan AP, PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indelsBioinformatics 2015 31(16):2745-47.10.1093/bioinformatics/btv19525851949 [Google Scholar] [CrossRef] [PubMed]
[31]. Capriotti E, Calabrese R, Fariselli P, Martelli PL, Altman RB, Casadio R, WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotationBMC Genomics 2013 14:01-07.10.1186/1471-2164-14-S3-S623819482 [Google Scholar] [CrossRef] [PubMed]
[32]. Pejaver V, Urresti J, Lugo-Martinez J, Pagel KA, Lin GN, Nam HJ, Inferring the molecular and phenotypic impact of amino acid variants with MutPred2Nat Commun 2020 11(1):591810.1038/s41467-020-19669-x33219223 [Google Scholar] [CrossRef] [PubMed]
[33]. Capriotti E, Fariselli P, Casadio R, I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structureNucleic Acids Res 2005 33(suppl_2):W306-10.10.1093/nar/gki37515980478 [Google Scholar] [CrossRef] [PubMed]
[34]. Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromoleculesNucleic Acids Res 2016 44(W1):W344-50.10.1093/nar/gkw40827166375 [Google Scholar] [CrossRef] [PubMed]
[35]. Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G, Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfacesBMC Bioinformatics 2010 11:54810.1186/1471-2105-11-54821059217 [Google Scholar] [CrossRef] [PubMed]
[36]. Sherry ST, Ward M, Sirotkin K, dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variationGenome Res 1999 9(8):677-79.10.1101/gr.9.8.67710447503 [Google Scholar] [CrossRef] [PubMed]
[37]. UniProt Consortium, UniProt: The Universal protein knowledge base in 2023Nucleic Acids Res 2023 51(D1):D523-31. [Google Scholar]
[38]. Irfan M, Iqbal T, Hashmi S, Ghani U, Bhatti A, Insilico prediction and functional analysis of nonsynonymous SNPs in human CTLA4 geneSci Rep 2022 12(1):2044110.1038/s41598-022-24699-036443461 [Google Scholar] [CrossRef] [PubMed]
[39]. Falahi S, Karaji AG, Koohyanizadeh F, Rezaiemanesh A, Salari F, A comprehensive in Silico analysis of the functional and structural impact of single nucleotide polymorphisms (SNPs) in the human IL-33 geneComput Biol Chem 2021 94:10756010.1016/j.compbiolchem.2021.10756034455166 [Google Scholar] [CrossRef] [PubMed]
[40]. Wang L, Yao Y, Si D, MMP-3 gene regulates the carcinogenesis and metabolic process of ovarian cancer, evidence from a Chinese population: Observational study and meta-analysisMedicine 2023 102(50):e3647110.1097/MD.000000000003647138115289 [Google Scholar] [CrossRef] [PubMed]
[41]. Durmanova V, Javor J, Parnicka Z, Minarik G, Ocenasova A, Vaseckova B, Impact of MMP2 rs243865 and MMP3 rs3025058 polymorphisms on clinical findings in Alzheimer’s disease patientsMediators Inflamm 2021 2021(1):557364210.1155/2021/557364233986628 [Google Scholar] [CrossRef] [PubMed]