Precision medicine is described as “an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person” [1]. Pharmacogenomics, the branch of science that combines Pharmacology (Science of Drugs) and Genomics (Study of genes and their functions) is a part of precision medicine. It looks into the effect of genes on a person’s response to a drug, to develop effective, safe medications and doses that are tailored to variations in a person’s genes [2].
The IHD is a condition that develops as a result of an imbalance between oxygen supply and demand of myocardium. It is relevant to note that of all the diseases affecting the cardiovascular system, IHD, according to statistics provided by the WHO in 2018, comes first in the list of 20 leading causes of death [3].
As per the global burden of diseases estimate (2010) cardiovascular diseases are the major cause of mortality and topping the list is IHD, with the age standardised death rate from CVD in India (272 per 100,000 population) being much higher than the global average (235 per 100,000 population) [4].
Pharmacotherapy plays an important role in management of patients with Coronary Artery Disease (CAD). There have been over 35 Randomised Clinical Trials (RCT), including more than 225,000 patients that studied DAPT in CAD, that includes combination of aspirin and an oral inhibitor of the platelet P2Y12 receptor for Adenosine di-phosphate (ADP) such as clopidogrel, prasugrel etc. Clopidogrel is considered the default P2Y12 inhibitor in patients with stable CAD treated with Percutaneous Coronary Intervention (PCI), those with indication to concomitant oral anticoagulation, as well as in ACS patients in whom ticagrelor or prasugrel are contraindicated. Ticagrelor or prasugrel is recommended in ACS patients unless drug-specific contraindications exist [5]. However, it is interesting to note that clopidogrel was prescribed despite DAPT guideline recommendations and RCT evidence suggesting superiority of the newer ADP receptor inhibitors [6].
Clopidogrel an important component of the regime is a prodrug, that must be converted to its active form to be effective. Rapidly metabolised in the liver, about 2% of clopidogrel finally gets oxidised to 2-oxo clopidogrel with the help of various enzymes of the cytochrome p450 family. Metabolism of clopidogrel is mostly carried out with the help of enzymes CYP2C19, CYP2B6, and CYP1A2 [7], with involvement of CYP3A4, CYP3A5, and CYP2C9 [8]. A 2-oxo clopidogrel is further metabolised to the active metabolite, with the help of aforementioned enzymes. The active metabolite which enters the blood stream, binds irreversibly to the P2Y purino receptor 12 on the surface of platelet cells, preventing ADP from binding to the receptor. Binding of ADP to the G protein coupled P2Y12 receptor is known to result in its activation, thereby stimulating adenylyl cyclase. Increase in intracellular cyclic Adenosine Monophosphate (cAMP) that results, activates calcium efflux pumps, and hence calcium build-up in the platelet. Eventually, this causes platelet activation, and later, aggregation promoting atherogenesis. Binding of clopidogrel to P2Y12 receptor inhibits this cascade, and hence prevent the progression of atherothrombotic processes [9]. Being a prodrug, a normal genetic make-up in whoever takes it is essential to ensure its efficacy, as several enzymes are involved in its metabolism. Numerous studies are conducted in this regard which report the presence of polymorphisms in genes which code for cytochrome family of enzymes that metabolise clopidogrel to its active form. CYP2C19 variants stand apart as they have been most commonly found to cause a variation in clopidogrel responsiveness [10].
Materials and Methods
The study was carried out as a cross-sectional study over a period of one year, between June 2018 to June 2019, among 60 patients who presented to Government Medical College, Thrissur, Kerala, India. Approval was obtained from the Institutional Research Committee (no. B6 8772/2016/MCTCR dated 30.11.2017).
Sample size calculation: The sample size was calculated using the formula, sample size=4pq/d^2, wherein, p=66%, q=100-p=34; d=20% of p; α error=1.96. Value of p was obtained from a previous study [13].
Inclusion criteria: Patients above 25 years of age, giving consent, diagnosed to have ACS, (by changes in their electrocardiogram and increase in serum cardiac troponin I levels) and started on DAPT for the first time, who have not been on any anti-thrombotics prior to presentation were included in the study.
Exclusion criteria: Patients with coagulation disorders and those with defects in platelet number or function were excluded.
Though there aren’t any genetic implications, clopidogrel naïve patients were selected with the aim of following them for future reference. About 2 mL of blood sample was taken from each patient into EDTA tubes about 24 hours after loading dose of clopidogrel was given upon presentation with ACS. This was then used for genetic analysis which was carried out at the laboratory under Jubilee Centre for Medical Research, Thrissur, Kerala, India.
CY2C19*2 Allele Testing
Heterozygosity or Homozygosity to CYP2C19 was identified by PCR-RFLP analysis of isolated Deoxyribonucleic Acid (DNA) using appropriate primers and restriction enzymes.
The procedure in brief is outlined below:
DNA isolation: Until the blood samples collected were used for DNA isolation, they were stored at -20°C as recommended by the isolation kit manufacturer. The procedure was performed using QIAmp DNA Blood Mini Kit (Catalogue Number: 51104, QIAGEN, Netherlands).
Isolated DNA was assessed qualitatively by agarose gel electrophoresis using Tris-Borate-EDTA (TBE) Buffer (prepared as 5X Standard stock; 54g Tris base, 22.5 gm boric acid and 20 mL 0.5M EDTA dissolved in 600 mL deionised water; final volume adjusted to 1 litre; 0.5X working solution prepared by dilution). Agar for the same was prepared in the concentration of 2% (0.8 gm agar in 40 mL, 0.5 X TBE buffer. Upon cooling, 2 μL Ethidium bromide (0.1) was added to the agar. This was then poured onto a casting tray with comb inserted to create wells in the gel. After the gel set, the comb was removed and samples with tracker dye was introduced into each well. A 6 μL of the sample with 2 μL of 6X loading dye (containing Glycerol, Bromophenol blue as front dye and Xylene cyanol FF, back dye) was loaded into wells along with a 50 bp ladder in the first well and water control in the last. Electrophoresis was performed at a current of 70 mA keeping the voltage constant. After gel run, the bands were visualised in GBoxChemi XRQ Imaging system with the help of the SYNGENE software.
Amplification of region of interest by polymerase chain reaction:CYP2C19 is located on exon 5 of chromosome 10 (Sequence from 94762681 to 94855547). The SNP rs4244285 corresponding to CYP2C19*2 allele is located at position 94781859 (change from G to A). Primers to obtain a short sequence of about 500 bp containing the nucleotide of interest, were designed with the help of National Centre for Biotechnology Information (NCBI) Primer Blast online tool. Parameters for primer design were a length of 15-25 bp, a melting temperature between 55-65°C and a GC content of 35-70%. Candidate primers were excluded if they had 3 or more G or C residues at the 3’ end, or if their annealing temperature differed by >3°C. Forward (5’TCAAAAGCAGGTATAAGTCTAGGAA3’) and Reverse (5’TCCTGACATCCTTATTGTTCAGAT-3’) primers obtained from Bioserve, were then used to create amplicon of length 667 base pairs. Upon receival, primers were reconstituted to 100 μmol as per instructions from the manufacturer. This was stored at -20°C as stock solution. Working solution prepared (10 μmol) by 1 in 10 dilutions as and when required. Reaction Mix (total volume 25 μL) that included forward and reverse primers (0.625 μL each), extracted DNA (3 μL), buffer (12.50 μL) and water (8.25 μL) were prepared in Eppendorf tubes and incubated in appropriately programmed thermal cycler for generation of amplicon [Table/Fig-1]. The amplicon obtained stored in Eppendorf tubes at -80°C until RFLP was done.
| No. | Step | Temperature | Duration | Number of cycles |
|---|
| 1. | Initial denaturation | 95°C | 5 Minutes | 1 |
| 2. | Denaturation | 94°C | 40 Seconds | 35 |
| 3. | Annealing | 60°C | 30 seconds |
| 4. | Extension | 72°C | 40 Seconds |
| 5. | Final extension | 72°C | 10 Minutes | 1 |
| 6. | Storage | 4°C | α | - |
Restriction digest of amplified products: The amplified products were subjected to restriction digestion to detect the presence of the allele. The enzyme selected in this regard was SmaI as seen from previous literature [14].
SmaI is known to produce blunt ends at 5’-CCCGGG-3’ sequence.
5’...C C C G G G... 3’
3’...G G G C C C... 5’
Confirmation of the same was carried out by virtual digestion of the amplicon with the help of online tool: restriction mapper version 3.
The enzyme was found to digest the normal allele into 2- one part with 277 bp and the other with 390 bp. The enzyme was found to have no effect on sequence carrying polymorphism.
Procedure for RFLP: Amplicon (10 μL), Buffer (2 μL), Water (7 μL), and SmaI (1 μL) were taken in 200 μL Eppendorf tubes.
The tubes were incubated at 30°C for 16 hours, after which the content of each tube was subjected to agarose gel electrophoresis using TBE buffer. Gel run was carried out as mentioned under DNA isolation. After gel run, the bands were visualised in GBoxChemi XRQ Imaging system with the help of the SYNGENE software.
Statistical Analysis
Statistical analysis carried out with the help of Epi Info software (CDC, USA). Quantitative variables were expressed as mean±Standard Deviation (SD). Categorical variables were expressed in frequency distribution. Genotype frequency was checked for deviation from Hardy Weinberg Equilibrium; being a cross-sectional study, the entire study population was considered for the same. Frequency of the variant allele was calculated from Hardy Weinberg equation.
Results
In the study, conducted among patients managed on clopidogrel, the frequency CYP2C19*2 allele was assessed to know the genotype of the study participants known to affect the efficacy of the drug. Study population comprised of males predominantly with 63% and only 37% of females. Highest frequency was in the age group between 61-65 years (22% in total). Patient demographics is elaborated in [Table/Fig-2].
| Patient demographics | Gender distribution | Total population |
|---|
| Males | Females |
|---|
| 38 (63%) | 22 (37%) | 60 (100%) |
|---|
| Age group (Mean Age) (years) | 37-77 (58) | 48-86 (63) | 37-86 (61) |
| H/o Diabetes mellitus (%) | 15 (39) | 13 (59) | 28 (47) |
| H/o Hypertension (%) | 13 (34) | 10 (45) | 23 (38) |
| H/o Dyslipidemia (%) | 3 (8) | 6 (27) | 9 (15) |
| H/o Smoking (%) | 25 (66) | 1 (5) | 26 (43) |
| H/o Alcohol consumption (%) | 25 (66) | 1 (5) | 26 (43) |
| H/o Similar illness in the family (%) | 16 (42) | 9 (41) | 25 (42) |
H/o: HIstory of
Genotype Frequency of CYP2C19*2 in the Study Population
Majority of the study population was found to carry the allele either in the heterozygous or homozygous state (62%). Heterozygosity for the allele was found in 42% individuals and homozygosity was found in 20%. A 681st position showed presence of the normal nucleotide in both the alleles in 38% of individuals [Table/Fig-3]. The interpretation of RFLP results is shown in [Table/Fig-4].
Gel electrophoresis showing restriction digest of amplicon.
Lane 1- 50 bp Ladder; Lanes 2 and 3- 2 bands at the 277 and 390 position- GG (Normal); Lanes 4 and 5: Single band at 667 position- AA (Homozygous for LOF Allele); Lanes 6 and 7: 3 bands corresponding to 667, 390 and 277 positions- AG (Heterozygous for Loss of Function allele; Lane 8: Water Control
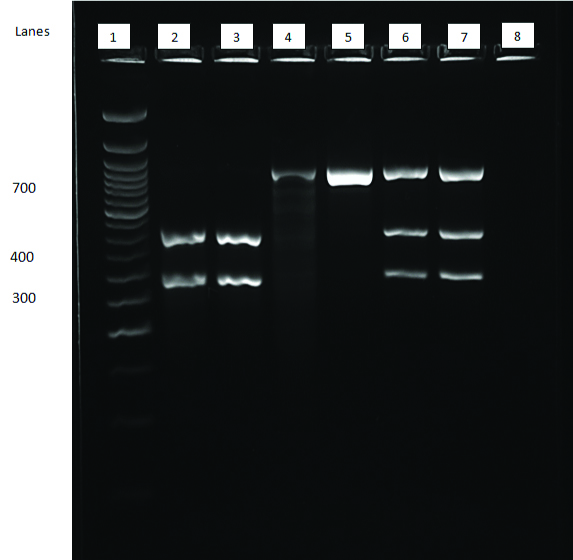
Interpretation of RFLP results.
| Observation | Inference | Result |
|---|
| 2 Bands: One corresponding to 277 and the other corresponding to 390 bp | Both alleles carry G at the 681st position of exon 5 | Normal (GG) |
| 3 Bands: One corresponding to 277, one band corresponding to 390 and another corresponding to 667 bp | One allele carries A and the other carries G at the 681st position of exon 5 of chromosome 10; Since one allele carries G, SmaI is able to cut it and as the other doesn’t, the enzyme is unable to cut it and therefore remains the original amplicon size of 667 | Heterozygous (AG) |
| 1 Band: Single band corresponding to 667 bp | As both the alleles carry A at the 681st position the enzyme is unable to cut it and hence only one band is observed. | Homozygous (AA) |
Genotype distribution was found to be in accordance with Hardy Weinberg Equilibrium (p<0.05 considered statistically significant and not in violation of Hardy Weinberg equilibrium). Allelic frequency in the study population *2 was found in 41% of population and *1 was found in 59% of population.
Discussion
The results of the study imply the presence of CYP2C19*2 genotype in majority (62%) of the study population. Being associated with therapy failure, the presence of the allele in the frequency observed seem to be an important aspect that require further probing in the setting of its use in prevention of atherothrombotic events. While the frequency of the abnormal genotype (AG/AA) was found to be higher than the normal genotype (GG), the frequency of the abnormal allele (*2) carrying the nucleotide A, was found to be lower than the normal allele (*1). Presence of heterozygotes in a higher number compared to homozygote variant could be the reason for this.
The in vitro demonstration of persisting platelet aggregation in previous studies among patients with the abnormal allele, even with administration of clopidogrel indicates likelihood of in vivo failure of desired effects with clopidogrel [14]. This suggests the chances of progression of the disease process or recurrence of similar episodes in future. To counter this, existing literature recommends classification of patients based on their genotype and subsequent adjustment of dosage or switching over to other antiplatelet drugs such as ticagrelor, prasugrel etc., in accordance with the Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and clopidogrel therapy [Table/Fig-5] [15,16].
CPIC (2013) antiplatelet therapy recommendations based on CYP2C19 status.
| Phenotype | Examples of diplotypes | Implications for clopidogrel | Therapeutic recommendations for clopidogrel in ACS/PCI |
|---|
| Normal metaboliser | *1/*1 | Normal platelet inhibition | Dose recommended by drug label |
| Intermediate metaboliser | *1/*2*1/*3*2/*17 | Reduced platelet inhibition; increased risk for adverse cardiovascular events | Alternative antiplatelet therapy recommended if no contraindication, e.g., prasugrel, ticagrelor |
| Poor metaboliser | *2/*2*2/*3*3/*3 | Significantly reduced platelet inhibition; increased risk for adverse cardiovascular events |
Generally, with respect to CYP2C19, individuals are classified as normal metabolisers if they are homozygous for the CYP2C19*1 allele (i.e., they are CYP2C19*1/CYP2C19*1), intermediate metabolisers if they have one CYP2C19*1 allele plus one variant allele (such as CYP2C19*2 or CYP2C19*3), and poor metabolisers if they carry two copies of a variant [15,16].
The CYP2C19*2 allele, along with the *3, *4, and *5 alleles, have been associated with decreased metabolic activity and have thus been termed LOF alleles. CYP2C19*2 allele is the most frequently occurring variant allele in Caucasian, African-American, and Asian populations. However, the allele frequency differs by racial group. CYP2C19*2 allele frequency in Asian populations (~30%) is significantly higher than that seen in Caucasians (~13%) and African-Americans (~18%). CYP2C19*3 allele occurs more frequently in Asian populations (~10%) compared with other racial groups (<1%) [17].
The CYP2C19*2 variant is the most common reason for poor metabolism of clopidogrel as well as other compounds like mephenytoin (an anticonvulsant), some antidepressants and some drugs used for ulcer conditions of various types [11]. rs4244285 (c.681G>A) is the defining polymorphism of CYP2C19*2 allele. It was previously referred to as CYP2C19m1. A synonymous G > A transition in exon 5 is the change observed; this creates an aberrant splice site which alters the mRNA reading frame, resulting in a truncated, non functional protein [18].
As with CYP2C19 variants, an ethnic variation in the distribution of CYP2C19*2 allele is also observed across the various ethnic groups around the world. A prospective cohort study from North India amongst 110 IHD patients showed that loss of function CYP2C19*2 was common in the Indian population as compared to other populations [12]. From the western part of the country, a study conducted in 2011 among 26 clopidogrel naïve patients with 102 healthy individuals ascertained that, polymorphisms in CYP2C19 resulting in loss of functional alleles does indeed play a role in reducing the activity of the enzyme and thus the action of clopidogrel as evidenced by the analysis of their platelet aggregation by turbidimetric method using ADP as an agonist [14].
In 2010-2011 a study carried out among 151 patients suffering from IHD in Puducherry concluded that genetic polymorphisms of CYP2C19 does influence the response to clopidogrel in patients with IHD of South Indian (Tamilian) descent with 71.1% of the population predicted to be poor or intermediate metabolisers of the drug [19]. Similar study in the year 2012 amongst 100 Cardiology outpatients of Meditrina Cardiac Centre, St. Thomas Hospital, Changanassery, Kerala, concluded that there is almost two times higher prevalence of CYP2C19*2 variant compared with other regions in the country [13].
A review by Swen JJ et al., reports that while healthcare professionals have successfully adopted the concept of pharmacogenomics and patients are willing to consent to participate in pharmacogenomic implementation studies, healthcare professionals do not frequently order or recommend a pharmacogenomic test [20]. Given the relevance of the studies of this nature, one cannot overlook this hurdle. Reasons for the same must be probed and adequate measures to overcome the same must be undertaken as it may lead to misdiagnosis and may be even life threatening for the patient when associated with diseases as in the current study.
Limitation(s)
The current study was carried out by PCR-RFLP method to look for the presence of only one of the polymorphisms associated with CYP2C19; this restricts the detection of other polymorphisms associated with the gene that may be present in the population as evidenced by discordant findings in genotype and phenotype analysis. Considering platelet aggregation studies with ADP as stimulant would aid in supplementing findings of genotype analysis as it permits assessment of the phenotype in vitro. In addition, better results may be obtained if the aggregation studies are conducted as a comparison between degree of platelet aggregation before initiation of therapy and after; or at different time points following initiation of therapy. The present study was carried out in a small group of patients that showed presence of the allele in significant numbers. This calls for the need to expand the study population and look for the allele in a larger group of individuals.
Future Recommendations
The presence of the allele in the magnitude as reported by this study, suggest that further studies be conducted which encompasses a wider population, for the following reasons: (i) The gene is found to be associated with metabolism of several other drugs; hence, the presence of the polymorphism puts patients at a higher risk of being hypo-responders or hyper-responders to the particular drug therapy; (ii) Assess the available Point of Care Testing devices or develop new ones, to look for inhibition of platelet aggregation in response to ADP, so that phenotype analysis may be performed easily in the absence of availability of genetic testing; (iii) For validation of test procedure; (iv) Despite being a field evolving at a rapid pace and having wide acceptance with significant application in management of various diseases, there is still an unmet need in validation and use of pharmacogenomics in routine clinical practice.
Conclusion(s)
The study indicates that CYP2C19*2 allele is present in the study population at a frequency comparable to the findings from studies conducted in India, confirming the incidence of the allele at a high frequency among people of Indian descent. The result from this study, as well as studies from the other parts of the country, suggests the need to ardently screen all patients to be initiated on clopidogrel therapy as the allele is associated with the risk of therapy failure that may be life threatening for the patient. Failure of therapy would invariably be associated with mortality and morbidity that is avoidable. In this regard, assessment of cost-effectiveness or cost utility of introduction of genetic testing prior to initiation of clopidogrel therapy compared to standard therapy in patients requiring antiplatelet regime may prove to be path breaking.
H/o: HIstory of