The UTI is one of the most common and frequent bacterial infection in both the community and hospital setting worldwide and it is an important health problem in all age groups. UTIs account for more than 7 million visits to doctor’s place and complicate over 1 million hospital admissions annually. Community Acquired (CA)-UTIs affect more than 150 million people annually while the prevalence of healthcare associated-UTIs ranges between 1.4- 5.1%. Members of the Enterobacteriaceae family are well-known for the cause of UTIs, predominantly Escherichia coli (E. coli) followed by Klebsiella and Proteus [1,2].
Materials and Methods
The present retrospective study included 207archived Klebsiella spp. isolates which were collected from UTI patients and maintained at the Department of Microbiology, Dr. ALM PG IBMS, University of Madras, Tamil Nadu, India, were included in the study and were stored and maintained at -20°C between December 2017 to January 2019. Retrospective analysis of these archived isolates was performed during Janurary 2020 to March 2020.
The isolates were revived in Macconkey agar and the colonies were subjected to gram staining and biochemical tests to confirm the Klebsiella spp.
Antimicrobial Susceptibility Testing
All the isolates were subsequently subjected to antimicrobial susceptibility testing by Kirby-Bauer disc diffusion method [10]. Extended Spectrum ß-Lactamase (ESBL) resistance was detected using ceftazidime (30 μg), aztreonam (30 μg), cefotaxime (30 μg), ceftriaxone (30 μg) discs for screening, followed by confirmatory test using ceftazidime (30 μg) and ceftazidime/clavulanic acid (10/5 μg) discs. The imipenem-EDTA Combined Disc Test (CDT) was performed to confirm carbapenem resistance among the study isolates, while AmpC lactamases resistance was detected using cefoxitin (30 μg) [11]. Resistance to fluoroquinolones was detected using discs of nalidixic acid (30 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), ofloxacin (5 μg), and levofloxacin (5 μg). Gentamicin (10 μg) and amikacin (30 μg) discs were used to detect resistance to aminoglycosides [11]. The isolates were also tested against other antibiotics like ampicillin (10 μg), piperacillin (100 μg), co-trimoxazole (1.25/23.75 μg), meropenem (10 μg), cefepime (30 μg), azithromycin (15 μg), fosfomycin (200 μg), tetracycline (30 μg) and chloramphenicol (30 μg). E.coli (American Type Culture Collection (ATCC) 25922) and Klebsiella (ATCC 700603) were used as control strains. The inhibition zones were measured, and results were interpreted according to the CLSI guidelines [11].
Detection of HMV Phenotype using String Test
The HMV phenotype was determined using a modified string test. Klebsiella isolates was cultured on MacConkey agar and the 24 hours culture of Klebsiella obtained were touched with an inoculation loop and then the loop was lifted from the agar surface. Strains were defined as HMV if there was formation of a viscous string longer than 5 mm [7].
Biofilm Detection Assays
The biofilm producing capacity of the Klebsiella isolates was determined by Congo Red Agar (CRA) method and tissue culture plate method. The qualitative detection of biofilm production was performed by CRA method. Briefly, the Klebsiella isolates were inoculated in the CRA medium, composed of brain heart infusion broth (37 gm/L), sucrose (5 gm/L), agar number 1 (10 gm/L) and Congo red dye (0.8 gm/L). Plates were incubated at 37°C for 24 to 48 hours aerobically. Black colonies with a dry crystalline consistency indicated biofilm production [Table/Fig-1].
Black crystalline biofilm production by standard control strain of E. coli in Congo Red Agar (CRA) medium.

Tissue culture plate method, which is a quantitative, gold-standard method for biofilm detection was performed for all the Klebsiella isolates. Briefly, 1 μL of overnight culture was inoculated into 100 μL of fresh Luria-Bertani (LB) broth in each well of a 96-well polystyrene plate. Negative control wells contained uninoculated sterile broth. After 5-24 hours of static incubation at 37°C, non-adherent bacteria were removed by gentle tapping and the wells were washed gently three times with 200 μL of Phosphate Buffer Saline (PBS) (pH 7.2). The bacteria were then stained with 200 μL of 0.1% crystal violet for one hour. The supernatant was discarded, and plates were washed with 200 μL of PBS to remove unattached cells. The biofilm-bound dye was then eluted with 200 μL of 70% acetic acid, and the optical density at 595 nm (OD595) was determined. The suitable positive control was used. Each assay was performed in triplicate. Based on the adherence capabilities, the strains were classified as given in [Table/Fig-2] [9].
Formula to interpretate biofilm production.
| Average OD value | Biofilm production |
|---|
| OD≤ODc* | Nil |
| ODc<OD≤2×ODc | Weak |
| 2ODc<OD≤4×ODc | Moderate |
| 4×ODc<OD | Strong |
*ODc-OD cut-off-Three standard deviations above the mean OD of the negative control
Statistical Analysis
The data obtained by quantitative biofilm assay was analysed by Student’s t-test using the online statistical software, GraphPad QuickCalcs. The results were expressed in the form of mean±SD (Standard Deviation). The p-values <0.05 were considered to be statistically significant.
Results
Bacterial strains: Among the 207 Klebsiella isolates, 196 (94.6%) isolates were Klebsiella pneumoniae subspecies pneumoniae, 2 (1.0%) isolates were Klebsiella oxytoca, 2 (1.0%) isolates were Klebsiella aerogenes and 7 (3.4%) belonged to Klebsiella pneumoniae subspecies ozaenae.
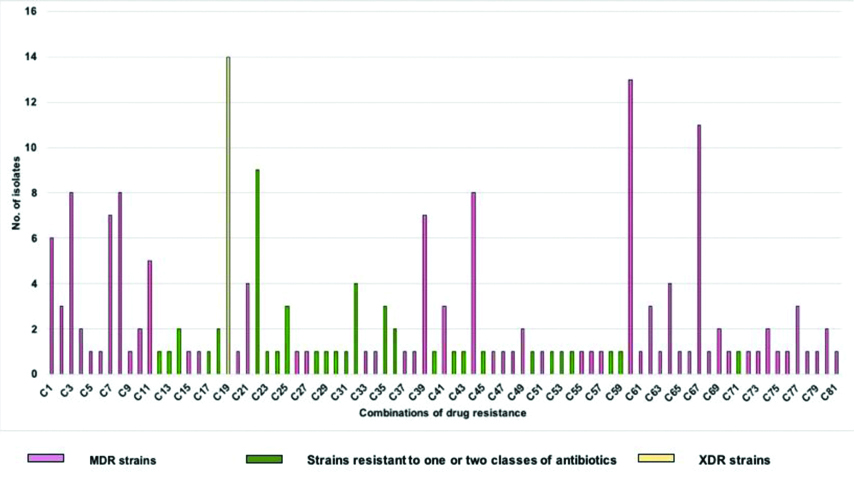
Drug resistance: Out of 207 isolates included in the study, 14 isolates (6.8%) were XDR; 141 isolates (68.1%) were MDR; 43 isolates (20.8%) were resistant to only one or two classes of antibiotic, while 9 isolates (4.3%) were totally susceptible. ESBL production was observed in 159 (76.8%) of the isolates. Amp-C lactamases were detected in 141 (68.1%) of the isolates. By CDT, carbapenem resistance was detected in 74 (35.7%) of the isolates.
Resistance to cotrimoxazole, cefepime and azithromycin was observed in 147 (71%), 159 (76.8%) and 165 (79.7%) of the Klebsiella isolates respectively. Highest sensitivity was observed for chloramphenicol 112 (54.1%), followed by fosfomycin 108 (52.2%). The [Table/Fig-3] summarises the antibiotic profile of Klebsiella isolates used in this study.
Antibiogram of Klebsiella isolates included in this study.
| Antibiotics | Concentration (μg) | Interpretation |
|---|
| Resistant n (%) | Intermediate n (%) | Susceptible n (%) |
|---|
| Cephalosporins |
| Cefoxitin- 2nd Gen | 30 | 141 (68.1) | 9 (4.3) | 57 (27.5) |
| Cefotaxime- 3rd Gen | 30 | 175 (84.5) | 13 (6.3) | 19 (9.2) |
| Ceftriaxone- 3rd Gen | 30 | 164 (79.2) | 9 (4.3) | 34 (16.4) |
| Ceftazidime- 3th Gen | 30 | 159 (76.8) | 9 (4.3) | 39 (18.8) |
| Cefepime- 4th Gen | 30 | 159 (76.8) | 5 (2.4) | 43 (20.8) |
| Carbapenems |
| Meropenem | 10 | 108 (52.2) | 22 (10.6) | 77 (37.2) |
| Imipenem | 10 | 74 (35.7) | 44 (21.3) | 89 (43.0) |
| Fluoroquinolones |
| Nalidixic acid- 1st Gen | 30 | 155 (74.9) | 4 (1.9) | 48 (23.2) |
| Ciprofloxacin- 2nd Gen | 5 | 157 (75.8) | 15 (7.2) | 35 (16.9) |
| Norfloxacin- 2nd Gen | 10 | 153 (73.9) | 5 (2.4) | 49 (23.8) |
| Ofloxacin- 2nd Gen | 5 | 146 (70.5) | 11 (5.3) | 50 (24.2) |
| Levofloxacin- 2nd Gen | 5 | 149 (72.0) | 7 (3.4) | 51 (24.7) |
| Aminoglycosides |
| Gentamycin | 10 | 102 (49.3) | 26 (12.6) | 79 (38.2) |
| Amikacin | 30 | 111 (53.6) | 19 (9.2) | 77 (37.2) |
| Penicillin’s |
| Ampicillin | 10 | 198 (95.7) | 4 (1.9) | 5 (2.4) |
| Piperacillin | 100 | 141 (68.1) | 29 (14.0) | 37 (17.9) |
| Other antibiotics |
| Ceftazidime/Clavulanic acid | 10.0/5.0 | 163 (78.74) | 0 | 44 (21.25) |
| Aztreonam | 30 | 149 (71.98) | 12 (5.79) | 46 (22.22) |
| Azithromycin | 15 | 165 (79.71) | 2 (0.96) | 40 (19.32) |
| Tetracycline | 30 | 127 (61.35) | 5 (2.41) | 75 (36.23) |
| Chloramphenicol | 30 | 86 (41.54) | 9 (4.34) | 112 (54.10) |
| Trimethoprim Sulfamethoxazole (Co-trimoxazole) | 1.25/23.75 | 147 (71.01) | 3 (1.44) | 57 (27.53) |
| Fosfomycin | 200 | 85 (41.06) | 14 (6.76) | 108 (52.17) |
Eighty-one different combinations of drug resistance patterns were observed among the 207 Klebsiella isolates included in the study (supplementary). Combination 19 (C19) was the most frequently observed combination with 14 (6.8%) isolates belonging to XDR category, followed by C60 and C67 with 13 (6.3%) and 11 (5.3%) of the isolates belonging to MDR category respectively. The details of other drug resistance combination patterns are shown in [Table/Fig-4].
Different combinations of drug resistance observed among the study isolates.
String test: Out of 207 isolates, only 2 (1.0%) isolates showed the HMV phenotype [Table/Fig-5].
Viscous string denoting Hyper-Mucoviscosity (HMV) of the Klebsiella strain.
Biofilm detection: In biofilm detection by CRA method, none of the isolates showed black crystalline formation.
In titre plate method, based on the ODc cut-off=0.048, 201 (97.1%) of the Klebsiella isolates were found to be strong biofilm producers (mean OD=0.88±1.06) and 6 (2.9%) of the isolates showed moderate biofilm formation (mean OD=0.17±0.02) [Table/Fig-6].
Biofilm production of the Klebsiella strain by titre plate method.
| OD value | Biofilm production level | Biofilm producers n (%) |
|---|
| ODc cut-off - 0.048 |
| >0.048 to ≤0.096 | Weak | 0/207 |
| >0.096 to ≤0.192 | Moderate | 6/207, (2.9%) |
| >0.192 | Strong | 201/207, (97.1%) |
Of the 201 strong biofilm producers, 13 (6.5%) were XDR strains (mean OD=0.43±0.18), 138 (68.6%) were MDR strains, 41 (20.4%) isolates were resistant to either one or two classes of antibiotics only (mean OD=0.44±0.20) and 9 (4.5%) of the Klebsiella isolates were susceptible to all the antibiotics (mean OD=0.43±0.19). Among the six moderate biofilm producing strains, 1 (16.7%) isolate was found to be XDR (mean OD=0.19±0) and 3 (50%) strains belonged to MDR category (mean OD=0.16±0.02) 2 (33.3%) isolates were resistant to either one or two classes of antibiotics only.
On analysing the OD values of XDR (mean OD=0.41±0.18) and MDR (mean OD=0.44±0.2) Klebsiella isolates, no significant variation was observed (p=0.5529). Similarly, there was a very little variation between the OD values of XDR and susceptible (mean OD=0.43±0.19) strains (p=0.8015).
Discussion
In the present study, the biofilm forming ability of the Klebsiella isolates with respect to antibiotic resistance was analysed, owing to the rapid emergence of MDR and XDR Klebsiella strains. In this study, majority of the Klebsiella isolates obtained from urine samples were identified as K. pnuemoniae. The result of this study was in line with other studies that have documented K. pnuemoniae as the predominant cause of UTI:3.8% of K. pnuemoniae isolates vs 0.7% of other Klebsiella spp. in Nepal [12]; 8.78% vs 5% in Pakistan [13] and 65.5% vs 34.5% in Ghana [14]. Mohammed MA et al., reported that K.pneumonia (16.3%) was the only Klebsiella spp. identified among 160 culture positive urine samples [15]. K. oxytoca as a cause of bacterial UTI is being frequently reported from other studies [14-17]. In the present study, 1% of the Klebsiella isolates were identified as K.oxytoca. In studies from Iraq and Zimbabwe, K.oxytoca was identified in 11% and 7% of the Klebsiella strains obtained from urine samples respectively [16,17]. In another study, K.oxytoca accounted for 38% of the Klebsiella-associated UTI [14]. In a study by Mohammed MA et al., K.pneumoniae subsp. pneumoniae (16.3%) was the most frequently detected Klebsiella species in urine samples, which was similar to present study. Also, in this study, K.oxytoca was identified in 2.5% of 160 bacterial species isolated from urine samples [15].
In this study, majority of the Klebsiella isolates were sensitive to chloramphenicol, followed by fosfomycin and imipenem. However, contrasting results have been documented in other studies. While Klebsiella strains showed maximum sensitivity to aztreonam, levofloxacin and piperacillin in a study from Turkey, another study from Russia found that all the urinary K. pneumoniae strains were sensitive to meropenem, amikacin and ciprofloxacin [18,19]. In line with present study findings, a study from North India found that the Klebsiella isolates were sensitive to fosfomycin and carbapenem group [20]. In South India, it was reported that the urinary K. pneumoniae isolates were sensitive to amikacin, ceftazidime and ceftriaxone [21].
High resistance was shown by the Klebsiella isolates used in this study to the commonly prescribed antibiotics like ampicillin, cotrimoxazole and ciprofloxacin. Similar results have been documented in other studies [21-23]. This could be due to the fact that these antibiotics have been used as first line drugs for UTI for many years, which has resulted in greater resistance in the community.
ESBL producers are more often implicated in UTIs, with prolonged duration of hospital stay, increased cost of hospitalisation and high mortality rate associated with ESBL-producing Klebsiella spp [24]. In present study, ESBL production was seen in 76.8% of the Klebsiella isolates which was very high when compared to other studies [21,25,26].
Carbapenems are the first choice of treatment for infections caused by Klebsiella strains producing ESBL and Amp-C enzymes [27-29]. The emergence of carbapenem-resistant K. pneumoniae has become a global threat. The results of this study showed that 35.7% of the Klebsiella isolates were carbapenem resistant, which was inconsistent with a previous study from South India [21].
String test revealed that only two isolates belonged to the HMV phenotype. This was in contrast to the results obtained by Vuotto C et al., wherein none of the Klebsiella spp isolates were classified as HMV, despite producing moist mucoid colonies [30].
It has been reported that the CRA method has very poor sensitivity in detecting biofilm when compared with the tissue culture plate method [8]. Similarly, in the present study none of the isolates were found to produce exopolysaccharide by CRA, while all the 207 isolates were found to be biofilm producers by tissue culture plate method. CRA method was not recommended for biofilm detection by Knobloch JK et al., in their study [31].
Biofilm producing organisms are considered to be intrinsically more resistant to antimicrobial agents. In the present study, 97.1% of the Klebsiella spp isolates were strong biofilm producers. However, no significant association was found between antibiotic resistance and biofilm forming ability, contradicting the results of previous reports [30-32]. In support of present study findings, Nirwati H et al., observed no association between K. pnuemoniae MDR strains and biofilm production based on statistical analysis [33].
This study emphasises on the judicious use of last line drugs and provides adequate knowledge about the current trends in susceptibility patterns of urinary Klebsiella spp. isolates and also confers in-depth phenotypic correlation between drug resisting and biofilm producing capacity among urinary Klebsiella spp. isolates.
Limitation(s)
The present study focused on the phenotypic correlation in association in depth without any molecular analysis and a detailed molecular level research has to be conducted further to understand insights of the biofilm-associated antimicrobial resistance.
Conclusion(s)
In this study, majority of the Klebsiella strains were resistant to most classes of the drugs and resistance profile was found to be high with ESBL production. In this study, there was high incidence of MDR Klebsiella spp and majority of MDR Klebsiella isolates were strong biofilm producers. Hence, biofilm may be one of the important factors for the drug resistance among the Klebsiella species causing UTI.