Sepsis has a major role in morbidity and mortality among critically ill patients even after the initiation of newer antibiotics. Sepsis outcome can be improved by early diagnosis [1]. Sepsis accounts for 2% of hospital admissions in developed countries. In case of critically ill patients, sepsis constitutes between 6 and 30%. Annual incidence of sepsis is found about 18 million [2]. Higher mortality rate was found to be associated with delay in the initiation of appropriate antimicrobial therapy [3].
Blood cultures are considered as the gold standard method for the diagnosis of Blood Stream Infections (BSI) [4]. Antimicrobial stewardship suggests that de-escalation of antimicrobial therapy should be done on the basis of culture reports and results in decreased antimicrobial exposure and cost savings [5]. De-escalation has an important role in antimicrobial stewardship programs [6]. Broad-spectrum antibiotics are the main stay of treatment. But irrational and inappropriate use of those leads to emergence of Multi Drug Resistance Gram Negative (MDRGNs). Appropriate usage of these can reduce the emergence of MDRGNs [7].
Clinical and Laboratory Standards Institute recommended tests give false negatives in case of coAmpc producers. To overcome this we can supplement ESBL tests with AmpC inhibitors such as boronic acid or cloxacillin performing clavulanate- or tazobactam based ESBL confirmatory tests with cefepime, which is not a substrate for AmpCs [8]. In standard laboratory practice, the time interval between the flagging of a positive blood culture and the reporting the isolate as an ESBL producer is three to four days. By testing ESBLs directly from positive blood culture bottles without performing subculture onto solid media, in order to decrease the amount of time required for the reporting of ESBLs showed high sensitivity and specificity [9].
Direct ESBL detection from positive blood culture bottles reduce turnaround time to 24 hours. By using aztreonam which acts as an inhibitor of AmpCs as well as substrate for ESBLs it can detect ESBLs in cases of coAmpc producers [10]. Therefore, the present study aids in faster detection of the ESBLs and thereby facilitates the early administration of appropriate antimicrobial therapy and also plays an important role in infection control and prevention.
Materials and Methods
A prospective study was conducted between October 2020-February 2021 at Bangalore Medical College and Research Institute, Bangalore, India. Ethical committee clearance was obtained from the institution (IEC Approval number: BMCRI/PS/1847/2020-21). Informed consent was obtained from the patient.
Sample size calculation: Based on previous study, ESBL prevalence was about 68% [11].
n=Z2pq/d2Where, z=1.96, P=68, q=32, d=9.6,
n=(1.96)2×68×32/(9.6)2=90Inclusion and Exclusion criteria: Positive blood culture bottles revealing gram-negative bacilli by gram stain were included. Positive blood culture bottles showing organisms other than gram-negative bacilli on gram staining and negative blood cultures were excluded.
All blood samples received for culture and sensitivity were processed by using BacT/ALERT 3D automated blood culture system according to manufacturer’s instructions. Positive blood cultures showing GNB were directly subjected to ESBL detection using CAZ, CTX discs with and without clavulanate and AO disc with and without clavulanate. For identification of the organism cultures were simultaneously subjected to Vitek-2 compact (Biomerieux) [12]. ESBL detection using CAZ/CTX with and without clavulanate disc diffusion method as recommended by CLSI performed on those culture isolates which were ESBL producers by direct ESBL method and also performed genotyping for those culture isolates.
Direct ESBL Detection by Disk Diffusion Method
All positive blood cultures were subjected to Gram stain. A total of 100 positive blood cultures showing gram-negative bacilli on Gram stain were taken for further testing. A suspension equivalent to a 0.5 McFarland standard was prepared with blood culture broth. Performed lawn culture using the suspension on Mueller-Hinton agar plates. ESBL detection was done by CLSI M 100 recommended inhibitory based methods by using CTX and CAZ discs (Himedia) with and without clavulanate. Another method of ESBL detection was done by using Aztreonam discs (Himedia) with and without clavulanate. Clavulanate solution was prepared using potassium clavulanate powder (Associate biotech) in 10 μgm/mL solution plates were incubated at 37°C for 24 hours. ESBL producers showed an inhibition zone size difference of >5 mm between the with and without clavulanate [10].
Genotyping was done for CTX-M, TEM, SHV genes. DNA extraction was done using boiling method. Master mix was procured from (Ampliqon) and PCR was carried out using thermal cycler. A reaction volume of 20 μL constitutes of DNA template, PCR buffer, MgCl, dNTPs, taq polymerase, forward and reverse primers with nuclease free water. Denaturation was carried out at 95°C for five minutes, constituting about 30 cycles of one minute duration and at 58°C and extension step at 72°C [13]. The primer sequences used are shown in [Table/Fig-1] [14]. Gel electrophoresis was done using 1% agarose gel.
Primers used for amplification of CTX-M, TEM, SHV genes.
| Primer | Primer sequence (5’-3’) | Product size (bp) |
|---|
| CTX-M gene Forward | TTTGCGATGCAGTACCAGTAA | 544 bp |
| CTX-M gene Reverse | CGTATATCGTTGGTGGTGCCATA | 544 bp |
| TEM gene Forward | ATAAAATTCTTGAAGACGAAA | 1011 bp |
| TEM gene Reverse | GACAGTTACCAATGCTTAATCA | 1011 bp |
| SHV gene Forward | GGGTAATTCTTATTTGTCGC | 930 bp |
| SHV gene Reverse | TTAGCGTTGCCAGTGCTC | 930 bp |
bp: Base pair
Statistical Analysis
Statistical analysis was done by using Microsoft Excel sheet. Descriptive statistics like percentage was done in the study.
Results
Out of 100 positive blood cultures showing gram negative bacilli on gram stain, 33 were ESBL producers and 67 were found to be non ESBLS by direct disc diffusion method. Age distribution of patients whose blood cultures showed GNB and ESBL producing GNBs is listed in [Table/Fig-2,3], respectively. The positive blood cultures (30%) and ESBLs (60.60%) were mainly found among neonates. ESBLs were predominantly seen among male patients as shown in [Table/Fig-4]. Organisms isolated from the positive blood cultures and ESBL producers among them are shown in [Table/Fig-5]. Acinetobacter baumanni were the predominant organism among the positive blood cultures while Klebsiella pneumoniae were predominant ESBL producers. ESBL detection by direct disc diffusion method and reference methods were performed on positive blood cultures showing GNB on Gram stain as shown in [Table/Fig-6]. Out of these 33 ESBL producers, 24 were found positive by both by CAZ/CTX with and without clavulanate disc diffusion method as recommended by CLSI and AO/CL disc diffusion method whereas six were found positive for ESBL production only by AO/CL disc diffusion method as shown by [Table/Fig-7] and three isolates were positive for ESBL production only by by CAZ/CTX with and without clavulanate disc diffusion method as recommended by CLSI as shown by [Table/Fig-8]. The 33 positive blood cultures containing ESBL producers as detected by direct disc diffusion method were subcultured and these culture isolates were subjected to ESBL detection and 27 of these culture isolates showed ESBL production by CAZ/CTX with and without clavulanate as recommended by CLSI. Turnaround time was found to be 24 hours for direct ESBL detection whereas it was found to be 48-72 hours for detection of ESBL producers from culture isolates.
Age distribution among patients showing positive blood cultures.
| Age | Positive blood cultures showing GNB (n=100) |
|---|
| 0-<30 days | 30 |
| 30 days-1 years | 15 |
| 1-10 years | 5 |
| 10-20 years | 10 |
| 20-30 years | 5 |
| 30-40 years | 10 |
| 40-50 years | 5 |
| 50-60 years | 10 |
| 60-70 years | 10 |
Age distribution among patients showing ESBL producers (n=33).
| Age | ESBLs, n (%) |
|---|
| 30 days | 20 (60.60) |
| 30 days-1 years | 10 (30.30) |
| 10-20 years | 3 (9.09) |
Sex distribution among patients showing GNB and ESBL producers from positive blood cultures.
| Gender | Positive blood cultures showing GNB (n=100) | ESBLs (n=33) |
|---|
| Male | 63 | 20 |
| Female | 37 | 13 |
ESBLs from positive blood cultures.
| Organisms | Number of isolates (n) | Number of ESBL isolates (n) |
|---|
| Acinetobacter baumanni | 35 | 5 |
| Klebsiella pneumoniae | 31 | 16 |
| Escherichia coli | 18 | 8 |
| Salmonella typhi | 6 | 0 |
| Acinetobacter lwoffi | 5 | 2 |
| Enterobacter aerogenes | 5 | 2 |
| Total | 100 | 33 |
Detection of ESBLs by test and reference methods.
| ESBL producers (n=33) | Test method | Reference method |
|---|
| Direct disc diffusion method | CAZ/CTX with and without clavulanate disc diffusion method as recommended by CLSI | PCR |
|---|
| Klebsiella pneumoniae (16) | 16 | 12 | 14 |
| Escherichia coli (8) | 8 | 7 | 7 |
| Acinetobacter baumanni (5) | 5 | 5 | 4 |
| Acinetobacter lwoffi (2) | 2 | 2 | 1 |
| Enterobacter aerogenes (2) | 2 | 1 | 2 |
| Total | 33 | 27 | 28 |
Direct detection of ESBLs by AO/CL Method.
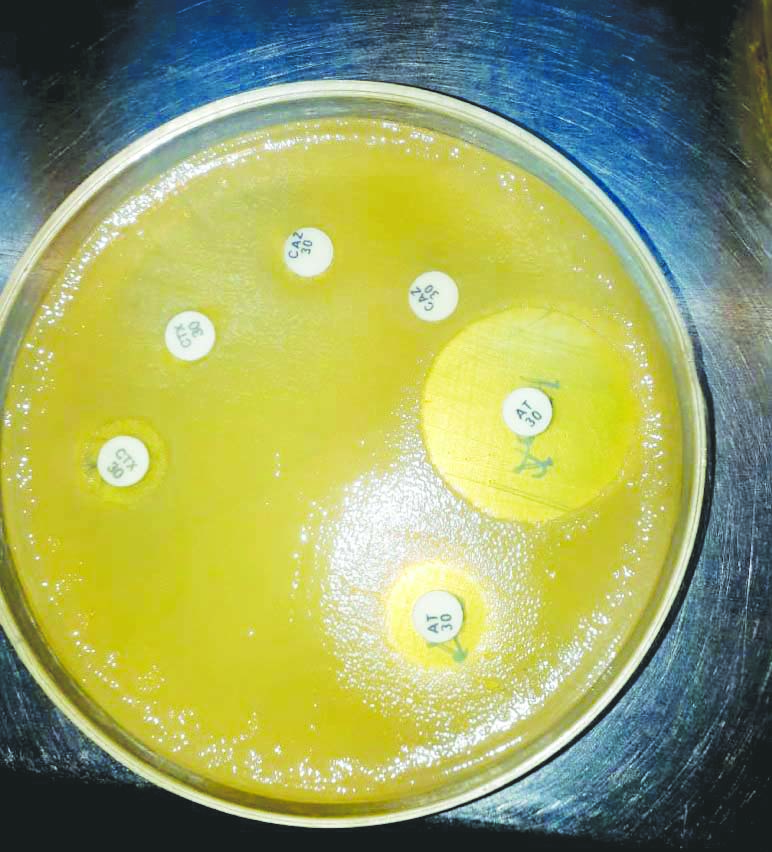
Detection of ESBLs by direct disc diffusion methods.
| ESBL producers (n=33) | Direct disc diffusion method |
|---|
| By CAZ/CTX with and without clavulanate disc diffusion method as recommended by CLSI only | By AO/CL disc diffusion method only | By both methods |
|---|
| Klebsiella pneumoniae (16) | 2 | 4 | 10 |
| Escherichia coli (8) | 0 | 1 | 7 |
| Acinetobacter baumanni (5) | 1 | 0 | 4 |
| Acinetobacter lwoffi (2) | 0 | 0 | 2 |
| Enterobacter aerogenes (2) | 0 | 1 | 1 |
| Total | 3 | 6 | 24 |
Out of 33 ESBLs, 28 had genes responsible for ESBL production such as TEM 10 (36%), CTX-M 10 (36%), TEM+CTX M 7 (25%), SHV 1 (3%) genes as detected by PCR as shown in [Table/Fig-9]. Gel electrophoresis pattern for TEM, CTX-M and SHV genes is shown in [Table/Fig-10,11 and 12]. In 05 (17.8%) isolates none of the target genes were found. Out of these five genotypically negative isolates, four were found positive for ESBL production by all the three phenotypic methods and one detected by AO/CL method alone. Out of six ESBL producers which were detected by only AO/CL method, five of them had genes responsible for ESBL production as shown in [Table/Fig-13].
Genotyping of ESBL isolates.
| Organisms (n=33) | Genotypic method (n=28) | Genes present |
|---|
| CTX M [n=10] (36%) | TEM (n=10) (36%) | CTX M+TEM (n=7) (25%) | SHV (n=1) (3%) |
|---|
| Klebsiella pneumoniae (16) | 14 | 5 | 6 | 3 | 0 |
| Escherichia coli (8) | 7 | 2 | 2 | 2 | 1 |
| Acinetobacter baumanni (5) | 4 | 2 | 1 | 1 | 0 |
| Enterobacter aerogenes (2) | 2 | 1 | 1 | 0 | 0 |
| Acinetobacter lwoffi (2) | 1 | 0 | 0 | 1 | 0 |
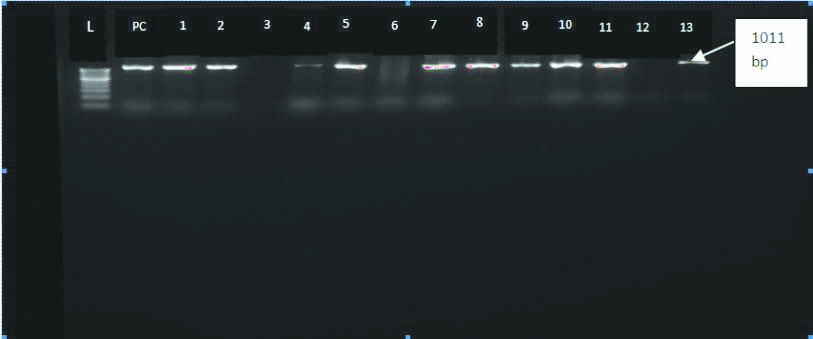
PCR for bla TEM GENE.
L-Ladder100-1000 bp; PC-Positive control TEM Gene positive isolates-1,2,4,5,7,8,9,10,11,13
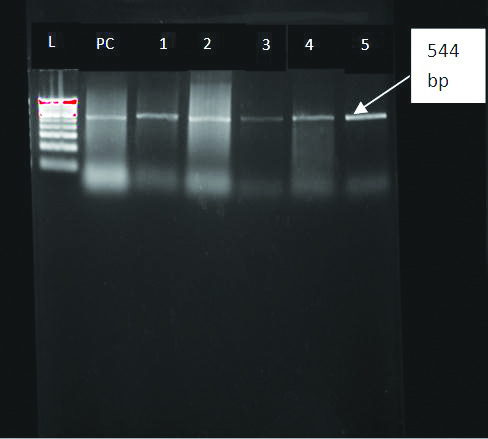
PCR for bla CTX-M GENE.
L-Ladder100-1000 bp; PC-Positive control CTX-M Gene positive isolates-1,2,3,4,5
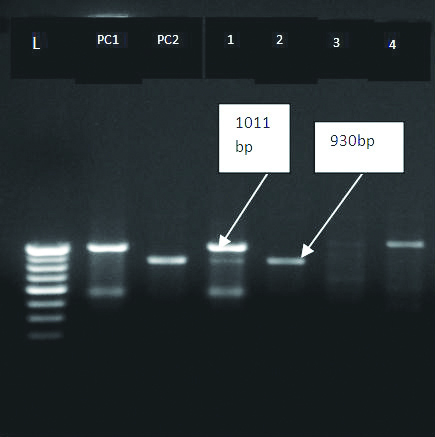
PCR for bla TEM and SHV GENES.
L-Ladder100-1000 bp; PC1-Positive control for TEM; PC2-Positive control for SHV TEMGene positive isolates-1,4; SHV Gene positive isolate-2
Detection of ESBLs by AO/CL and PCR.
| ESBL producers (n=6) | By AO/CL disc diffusion method only (n=6) | PCR (n=5) |
|---|
| Klebsiella pneumonia (4) | 4 | CTX-M-2 TEM-1 |
| Escherichia coli (1) | 1 | CTX-M+TEM-1 |
| Enterobacter aerogenes (1) | 1 | TEM-1 |
| Total | 6 | 5 |
Discussion
Rapid detection of ESBL producing bacteria is important to choose appropriate antibiotics for treatment. It plays a significant role in antibiotic stewardship and infection control. With this information clinician can either continue with broad spectrum antibiotics started as empirical treatment or step up with combination of β-lactam- β-lactamase inhibitors or carbapenems as justified with clinical condition. This study was taken up to detect ESBL producers directly from positive blood culture bottles hence shortening the delay; using Aztreonam disc with and without clavulanic acid was assessed along with CAZ/CTX with and without clavulanate as recommended by CLSI.
In the present study, ESBL producers from positive blood culture broth constituted 33% which is comparable with the studies conducted by Doret L et al., Bianco G et al., and Navon-Venezia S et al., [15-17]. Out of 33 (33%) ESBL isolates, 26 (78.78%) belongs to family Enterobacteriacea and remaining 7 (21.21%) belonging to nonfermenters. Among Enterobacteriacea family 16 (48.5%) of the ESBL isolates were of K. pneumonia followed by E. coli.
Direct ESBL detection from positive blood cultures using CAZ/CTX with and without clavulanate disc by CLSI guidelines and by AO/CL disc diffusion method detected 90% of ESBLs. A study conducted by Thomson GK et al., using improved method from the culture isolates also shown similar results shown by present study [8]. The advantage of this study is direct detection of ESBL producers from positive blood culture bottles showing GNB on Gram stain reduced the turnaround time for reporting of ESBL producers to 24 hours from obtaining positive blood culture which in contrast to more than 48-72 hours when ESBL producers are detected from culture isolates as done by Thomson GK et al., [8].
By using CAZ/CTX 81% of ESBLs were directly detected from positive blood cultures. This study detected 33% of ESBLs by phenotypic method from positive blood cultures. Similar results were shown by studies conducted Poulou A et al., Kazemian H et al., and Jabeen K et al., but performed on the culture isolates instead of using positive blood cultures [18-20]. In the present study, it was observed that ESBL detection from positive blood cultures by direct method (CAZ/CTX with and without clavulanate) were showing comparable results with ESBLs producers from culture isolates of those positive blood cultures by CLSI recommended method.
A 28% ESBLs showed positive for the ESBL target genes such as CTX-M, TEM, SHV which is similar to the study conducted by Krishnamurthy V et al., [14]. Present study showed 10 TEM (36%), 10 CTX-M (36%), seven TEM+CTX M(25%), one SHV (3%) genes. A 5 (15.15%) of ESBLs detected by phenotypic method didn’t possess any of the gene targets tested in this study. It may be due to the presence of ESBL genes other than the ones detected in this study.
Therefore, direct ESBL testing from positive blood cultures helps in early reporting of ESBLs which inturn guides the treating physician in facilitating rapid initiation of appropriate treatment. The time delay associated with CLSI standard methods accounts for 48-72 hours which constitutes sub culturing onto a solid culture medium, ESBL screening and confirmatory ESBL test. The present study showed that direct ESBL testing from positive blood culture bottles is easy to perform and yields results within 24 hours reducing time delay. Apart from that by using Aztreonam discs for ESBL detection shows additional advantage. Aztreonam is a substrate for ESBL as well as inhibitor of Ampc, it will increase the detection rate in case of CoAmpc producers.
Limitation(s)
Detection of ESBL producers were carried from 100 positive blood cultures showing GNB on gram stain. Larger sample size should be included in further studies in order to determine efficiency of this method.
Conclusion(s)
Direct detection of ESBL producers from positive blood culture specimens by using AO/CL disc is preferred because of reduced turnaround time and also an inexpensive test requiring only one substrate.