Introduction
Urinary Tract Infections (UTI) are one of the commonest conditions for which people seek medical care with an estimated 150 million episodes per annum worldwide. An unprecedented upsurge in the rate of development of antimicrobial resistance has reduced the therapeutic options leading to increased morbidity, prolonged hospital stays, development of complications. Majority of these infections are attributable to Gram negative bacteria which have now acquired resistance to almost all classes of antibiotics.
Aim
To analyse the plasmid-mediated drug resistance and characterise the major plasmid families that are in circulation.
Materials and Methods
A cross-sectional study comprising of a total of 95 non consecutive multidrug-resistant gram-negative bacterial isolates were subjected to Plasmid based replicon typing from January 2017 to June 2018. The 18 major replicons were divided in five multiplex and three uniplex Polymerase Chain Reaction (PCR) formats and the samples were subjected for plasmid characterisation and further sequencing of the plasmid Deoxyribonucleic Acid (DNA). The data obtained was analysed by Microsoft Excel software.
Results
Escherichia coli, accounted for maximum n=51 (53.7%), Klebsiella pneumoniae n=19 (20%), Citrobacter sp n=11 (11.6%), miscellaneous gram negative n=14 (14.7%) The isolates exhibited a high degree of resistance to almost all tested antibiotics, sparing a few like Fosfomycin, Chloramphenicol, Imipenem, Amikacin. A total of 154 different plasmid families were detected from the 95 isolates. FIB replicon (24%), FIA (21%), F, W (20%), FIC, B/O (14%), Y (12%), I1 replicon (10.5%) were the major plasmid families detected in the present study.
Conclusion
Many isolates exhibited the presence of more than one Incompatibility (Inc.) group plasmids, conferring multidrug resistance to the isolates. The study highlights the need for further research to study the association between plasmid families and their respective antibiotic resistance profiles for a given geographical niche and the need to devise further methods to target these epidemic plasmids.
Conjugation, Polymerase chain reaction, Replicon typing, Urinary tract infection
Introduction
Garret Hardin’s famous essay “In a world that is limited, ruin is the destination towards which all men rush” can aptly be put in the context of the ever increasing menace of antimicrobial resistance [1]. According to the CDC, 2.8 million antibiotic-resistant infections occur in the US each year, and more than 35,000 people die as a direct consequence of these infections [2]. Globally, World Health Organisation (WHO) estimates deaths due to drug-resistant organisms to be around 10 million annually by 2050 and may lead up to 24 million people into extreme poverty by end of this decade [3]. The era of discovery of antibiotics had ushered in a ray of hope of eliminating the bacterial diseases from the face of the world, but the hopes were shattered sooner than expected with the emergence of bacterial strains with resistance to these therapeutic agents.
A major contributor to the acquisition and subsequent horizontal transfer of resistance are the presence of extra chromosomal heritable determinants known as Plasmids. The role of plasmids in development of antibiotic resistance was first studied in Japan when it was found that susceptible strains were converting into multidrug-resistant ones, not as a result of mutations under selection pressure but by the acquisition of certain resistance determinant factors [4]. Both intergeneric and intrageneric transfer of resistance has well been documented in several studies [5-7]. Plasmids have been classified into various classes based on their replication controls and are termed as Incompatibility (Inc) groups. Plasmids which possess same replication control are incompatible whilst plasmids with different replication controls are termed as compatible plasmids and can co-exist within the same bacterium [8-10].
Bacteria have developed resistance to all classes of antibiotics including the ones which are often referred to as last resort antibiotics. The occurrence of resistance to Penicillins, Cephalosporins, Fluoroquinolones, Folate synthase Inhibitors, Carbapenems, Polymyxins are all known to exist in Plasmids and the list keeps on increasing day by day [11-14].
Until 2011, there was no clear cut definition of what comprised a Multidrug Resistant Organism (MDRO) in the context of bacteria. European Centre for Disease Prevention and Control (ECDC) and (CDC) defined it as an organism non susceptible to ≥1 anti-microbial agent in ≥3 antimicrobial categories [15]. They also defined Extensive Drug Resistance (XDR) and Pan Drug Resistance (PDR) as non susceptible to ≥1 agent in all but ≤2 antimicrobial categories and non susceptible to all antimicrobial agents respectively [15,16]. This study aimed to evaluate the presence of these major plasmid families in circulation amongst gram negative MDROs in our geographical niche and attempt to fulfill the gaps in knowledge about increasing antibiotic resistance in India.
Materials and Methods
This cross-sectional study was carried out at Armed Forces Medical College (AFMC), Solarpur, Pune, Maharashtra, India. The study was carried out over a period of 18 months from January 2017 to June 2018. The study was duly approved by Institutional Ethics Committee (AFMC, Solarpur, Pune, IEC meet held in 2015) and written informed consent was obtained from the patients. The main objectives of the study was to isolate, identify and perform antibiotic susceptibility testing on all gram negative isolates from UTI cases in our centre followed by analysing plasmid mediated drug resistance by performing molecular characterisation of these plasmids.
Sample size calculation: The culture positivity rate for urine sample was taken to be 15%. The sample size had been worked out based on the presumption of ~41% drug resistance in isolates where the power of the study is α=0.05 (CI=95%, d=10%) [17-19]. The total number of urine samples processed in order to obtain these 95 MDROs was 1500. MDRO is defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [15].
Inclusion criteria: Consecutive, non repeat, significant growth >=105 CFU/mL positive multi drug resistant gram negative bacterial isolates obtained from both outpatients and inpatients departments. All patients were clinically suspected to have UTI by the treating doctor.
Exclusion criteria: Urine cultures exhibiting Gram positive bacteria, Candida, Mixed growth obtained upon culture were excluded from the study and any growth <105 CFU/mL was considered non-significant growth and excluded from the study.
Sample collection: Mid-stream clean catch urine samples from suspected UTI cases were included in the study. For catheterised patients, urine aspirate was drawn aseptically from the catheter.
Microbiological Methods
Isolation and identification and antibiotic susceptibility testing of the samples n=95 were performed as per standard microbiological methods using semi-quantitative culture technique on Cysteine Lactose Electrolyte Deficient medium (CLED agar) (Hi-Media, India). Further identification and susceptibility was performed using automated Vitek2 Compact SL (Bio-Merieux Inc, SA Marcy-l’Etiole, France). Additional testing of antibiotics, not in the panel of Vitek2 was performed by Kirby Bauer disk diffusion method on Mueller-Hinton agar (Hi-Media, India) and results obtained were interpreted as per The Clinical and Laboratory Standards Institute (CLSI) 2017 and 2018 M-100 interpretive criteria [20].
Plasmid Extraction and Analysis
Plasmid DNA was extracted by miniprep spin column method using Hi Pur ATM Plasmid DNA Miniprep Purification Kit (Hi Media Labs, India) as per manufacturer’s instructions. The Plasmid DNA was preserved at -80°C until further analysis.
PCR based Replicon typing (PBRT): The PBRT system was initially developed and further validated by Dr. Alessandra Carattoli, at the Italian National Institute of health, Rome which comprises of 18 major plasmid incompatibility groups and replicates genes which were identified and subsequently sequenced and are known to be circulating amongst gram negative bacteria [21-23]. The replicons used were HI1, HI2, I1, X, L/M, N, FIA, FIB, W, Y, P, FIC, A/C, T, FIIS, FrepB, K, B/O. All primer sequences were procured from Sigma Aldrich, India [Table/Fig-1]. Primers used in the study were divided into five Multiplex (M1-M5) and three uniplex PCR (S1-S3) on the basis of their amplicon size to avoid overlapping of amplicon bands. Amplicon (Amp) sizes ranged from 139 bp-785 base pairs [22,24,25].
Sequences of primers used in the study.
| Plasmid family nomenclature | Sequence 5'-3' [21] | Amplicon size (bp) |
|---|
| HI1 FW | GGA GCG ATG GAT TAC TTC AGT AC | 471 |
| HI1 RV | TGC CGT TTC ACC TCG TGA GTA | |
| HI2 FW | TTT CTC CTG AGT CAC CTG TTA ACA C | 644 |
| HI2 RV | GGC TCA CTA CCG TTG TCA TCC T | |
| I1 FW | CGA AAG CCG GAC GGC AGA A | 139 |
| I1 RV | TCG TCG TTC CGC CAA GTT CGT | |
| X FW | AAC CTT AGA GGC TAT TTA AGT TGC TGA T | 376 |
| X RV | TGA GAG TCA ATT TTT ATC TCA TGT TTT AGC | |
| L/M FW | GGA TGA AAA CTA TCA GCA TCT GAA G | 785 |
| L/M RV | CTG CAG GGG CGA TTC TTT AGG | |
| N FW | GTC TAA CGA GCT TAC CGA AG | 559 |
| N RV | GTT TCA ACT CTG CCA AGT TC | |
| FIA FW | CCA TGC TGG TTC TAG AGA AGG TG | 462 |
| FIA RV | GTA TAT CCT TAC TGG CTT CCG CAG | |
| FIB FW | GGA GTT CTG ACA CAC GAT TTT CTG | 702 |
| FIB RV | CTC CCG TCG CTT CAG GGC ATT | |
| W FW | CCT AAG AAC AAC AAA GCC CCC G | 242 |
| W RV | GGT GCG CGG CAT AGA ACC GT | |
| Y FW | AAT TCA AAC AAC ACT GTG CAG CCT G | 765 |
| Y RV | GCG AGA ATG GAC GAT TAC AAA ACT TT | |
| P FW | CTA TGG CCC TGC AAA CGC GCC AGA AA | 534 |
| P RV | TCA CGC GCC AGG GCG CAG CC | |
| FIC FW | GTG AAC TGG CAG ATG AGG AAG G | 262 |
| FIC RV | TTC TCC TCG TCG CCA AAC TAG AT | |
| A/C FW | GAG AAC CAA AGA CAA AGA CCT GGA | 465 |
| A/C RV | ACG ACA AAC CTG AAT TGC CTC CTT | |
| T FW | TTG GCC TGT TTG TGC CTA AAC CAT | 750 |
| T RV | CGT TGA TTA CAC TTA GCT TTG GAC | |
| FIIS FW | CTG TCG TAA GCT GAT GGC | 270 |
| FIIS RV | CTC TGC CAC AAA CTT CAG C | |
| FrepB FW | TGA TCG TTT AAG GAA TTT TG | 270 |
| FrepB RV | GAA GAT CAG TCA CAC CAT CC | |
| K/B FW | GCG GTC CGG AAA GCC AGA AAA C | 160 |
| K RV | TCT TTC ACG AGC CCG CCA AA | |
| B/O RV | TCT GCG TTC CGC CAA GTT CGA | 159 |
All PCR amplifications were carried out in Gene Amp PCR system 97°C (Applied Biosystems) with the following protocol; one cycle of denaturation at 94°C for five minutes, 30 cycles of Denaturation at 94°C for one minutes, Annealing at 55°C for 30 seconds, Elongation at 72°C for one minute followed by final extension program of one cycle at 72°C for 5 minutes. In Uniplex one PCR carrying F rep plasmid the annealing temperature was kept at 50°C, rest of the PCR protocols were kept same. The post PCR products were electrophoresed on 1% agarose gel and viewed under Ultraviolet (UV) transilluminator.
Sequencing study: Pure LinkTM Quick Gel Extraction, Invitrogen by Life technologies was used as per manufacturer’s instructions for DNA purification and extraction. ABI 3730 XL Analyser (Applied Biosystems, CA) based on Sanger’s dideoxy DNA Capillary sequencing was used. The sequence was analysed using ABI Portal Resources for Indiana Science and Mathematics (PRISM) SeqScape version2.0 software.
Statistical Analysis
The data were entered in Microsoft Excel and analysed. Frequency (n) and percentages (%) were analysed for the collected data. The sequence got in all the samples was analysed using ABI (PRISM) SeqScape vers.
Results
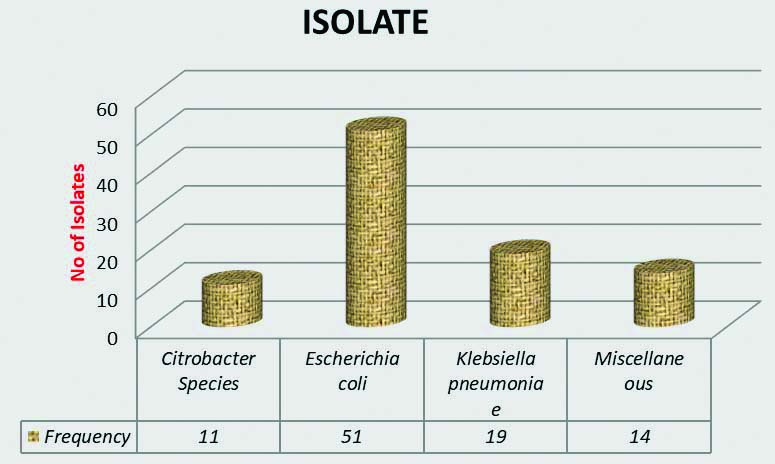
The total number of urine samples processed was 1500, out of which 95 culture positive MDROs were selected for the study. Majority of the present study population was in the age group of 15-45 years n=57 (60%), followed by above 45 years n=30 (31.6%) and below 15 years of age n=8 (8.4%). Frequency distribution of gender profile had no major difference with almost equal male n=49 (51.6%) to female ratio n=46 (49.4%). Outpatients accounted for n=30 (31.6%) of the study size whilst inpatients were n=65 (68.4%). Amongst frequency distribution of isolates [Table/Fig-2]; Escherichia coli n=51 (53.7%), Klebsiella pneumoniae n=19 (20%), Citrobacter sp n=11 (11.6%), Miscellaneous n=14 (14.7%) were the other gram negative isolates obtained. Amongst the Miscellaneous group Pseudomonas spp n=6 (6.2%), Proteus spp n=4 (4.2%), and one isolates each of Myroides odoratimimus and Serratia marcescens, Enterobacter aerogenes, and Stenotrophomonas maltophila were obtained. Analysis of the antibiogram of the MDR isolates [Table/Fig-3] showed a very high resistance to Ampicillin, Amoxycillin-clavulanate and third generation Cephalosporins. However, Fosfomycin, Chloramphenicol were the only few antibiotics to which resistance was found to be relatively low.
Bar graph depicting frequency distribution of Isolates (n=95).
Resistance pattern of the isolates for the tested antibiotics (n=95).
| Antibiotic | Percentage resistance n (%) |
|---|
| AMP | 84 (88.4) |
| PI | 82 (86.3) |
| CPM | 54 (57.5) |
| CTX | 68 (71.5) |
| CTR | 53 (55.7) |
| CAZ | 45 (47.8) |
| GEN | 23 (24.7) |
| AK | 18 (18.9) |
| SAM | 76 (80.6) |
| PIT | 48 (50.5) |
| CIP | 63 (66.3) |
| LE | 63 (66.3) |
| OF | 44 (46.9) |
| NOR | 68 (71.5) |
| ETP | 22 (23.2) |
| IMP | 15 (16.1) |
| MRP | 37 (38.6) |
| COT | 53 (56.4) |
| NIT | 48 (50.5) |
| AMC | 86 (90.5) |
| C | 4 (4.2) |
| AZT | 73 (77.0) |
| NAL | 83 (87.3) |
| FOS | 9 (9.5) |
AMP: Ampicillin; PI: Piperacillin; CPM: Cefepime; CTX: Cefotaxim; CTR: Ceftriaxone; CAZ: Ceftazidime; GEN: Gentamicin; AK: Amikacin; SAM: Ampicillin-sulbactam; PIT: Piperacillin-tazobactam; CIP: Ciprofloxacin; LE: Levofloxacin; OF: Ofloxacin; NOR: Norfloxacin; ETP: Ertapenem; IMP: Imipenem; MRP: Meropenem; COT: Cotrimoxazole; NIT: Nitrofurantoin; AMC: Amoxicillin clavulanate; C: Chloramphenicol; AZT: Aztreonam; NAL: Nalidixic acid; FOS: Fosfomycin
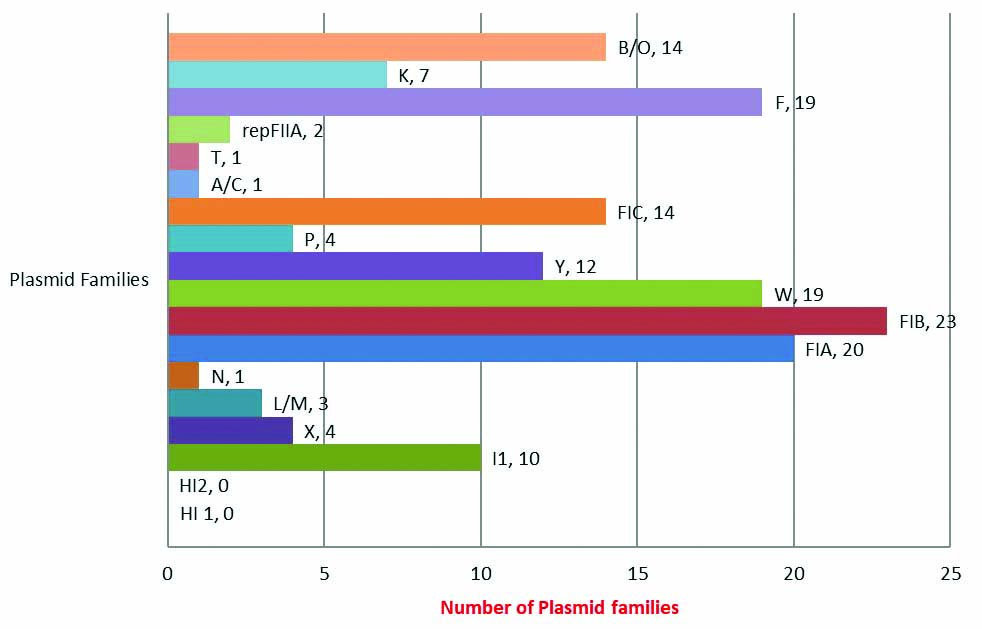
A total of 154 Plasmid replicon families were detected from the total isolates (n=95) confirming presence of multiple plasmid families within a single MDR isolate. Of the 18 gene targets, the indepth study found the presence of 16 plasmid rep families. FIB replicon was the commonest plasmid family detected in 23 out of 95 (24.2%) samples [Table/Fig-4]. Of the 16 Plasmid families, only eight products gave sequences that were subsequently [Table/Fig-5] converted in FASTA files and were analysed using the NCBI BLAST software program (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Bar graph depicting frequency distribution of plasmid families (n=154).
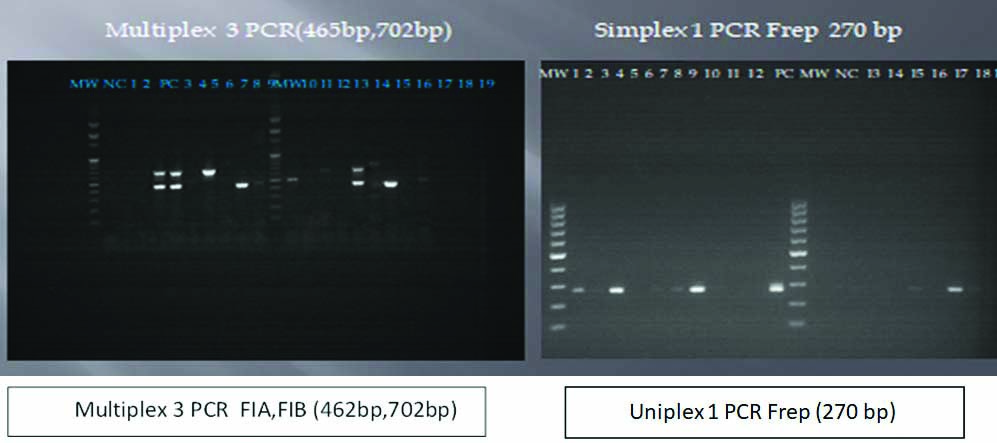
MW-100 bp molecular weight ladder.
PC: Positive control; NC: Negative control; Lane 1- 19: Plasmid DNA (from left to right: Lane 4 FIB replicon (702 bp), Lane 7 FIA replicon (462 bp), Lane 13 containing both FIA and FIB replicons, Uniplex 1 PCR Frep plasmids seen in Lanes 1,3,9,15 and 17)
Discussion
The UTIs are one of the commonest presentations in any hospital setting and various studies conducted worldwide have suggested prevalence of UTI being much higher in females as compared to males [26-28]. The study had a slight male preponderance with 51.6% of males as compared to 48.4 % females. Most of the isolates were obtained from the age group of 15-45 years, which accounted for 60% followed by >45 year of age (31.6%). Only 8.4% of the samples were from the paediatric age group. This variation in the gender distribution among the adult population is likely due to our patient pool comprising mainly of serving soldiers, veterans and their families and dependents. The results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) study concluded similar findings in which the authors found that family Enterobacteriaceae comprised 86.0% of all the isolates out of which Escherichia coli accounted for 56.5% and Klebsiella pneumoniae was found in 13.8%, occuring as the two most common species isolated [29]. Antimicrobial susceptibility data commensurated with findings of various multi centric studies [16,30-34]. The overall high prevalence of resistance to oral antimicrobials is also explained by the indiscriminate use of these antibiotics in the present setting. Amongst the injectable agents, the susceptibility rates were better than the oral as their usage is somewhat regulated and antimicrobial stewardship is more in these cases. Cross-resistance or Co-Selection may also be a reason for higher resistance amongst the studied uropathogens. In various studies of Caratolli A, the presence of IncFII, IncA/C, IncL/M, and IncI1 plasmids showed the highest occurrence among typed resistance plasmids [10,21,22,35]. Presence of these plasmids can be rightfully termed as Epidemic plasmids as they accounted for majority in these isolates and the resistance genes they harbor. Association studies between Plasmid families and their respective antibiotic resistance profiles were attempted but could not be interpreted by Chi-square test because of very small frequencies. The significant families that were associated most commonly with resistance were found to be located in FIB, FIA [Table/Fig-5], Frep, W, FIC, I1 and B/O replicons in order of decreasing frequency. However, since the bacteria has multiple resistance mechanisms to exhibit for the same antibiotic, finding a definite plasmid coding for a particular antibiotic resistance would not be entirely correct and more studies with larger sample sizes would be required to have an accurate correlation between the same. The information provided here further adds to the understanding of the mechanisms of drug resistance in this specific region and their evolutionary relationship with other parts of world [23].
Limitation(s)
A larger sample size may have delineated the association between the Plasmid families and antibiotic resistance. The study did not characterise Chromosome mediated drug resistance.
Conclusion(s)
Having an insight into the major plasmid families circulating in the present setting can help us understand the resistance profiles and also help us in specifically targeting those. This would go a long way in our efforts to implement antimicrobial stewardship in a more targeted and efficient manner. The study aimed at understanding the transferable drug resistance amongst gram negative bacterial isolates and molecular characterisation of the plasmids that were found in the study. The study highlights the importance to know the plasmid families that are prevalent in a given geographical niche and understand the resistance genes that they carry and devise further methods to target such epidemic plasmids using novel techniques.
AMP: Ampicillin; PI: Piperacillin; CPM: Cefepime; CTX: Cefotaxim; CTR: Ceftriaxone; CAZ: Ceftazidime; GEN: Gentamicin; AK: Amikacin; SAM: Ampicillin-sulbactam; PIT: Piperacillin-tazobactam; CIP: Ciprofloxacin; LE: Levofloxacin; OF: Ofloxacin; NOR: Norfloxacin; ETP: Ertapenem; IMP: Imipenem; MRP: Meropenem; COT: Cotrimoxazole; NIT: Nitrofurantoin; AMC: Amoxicillin clavulanate; C: Chloramphenicol; AZT: Aztreonam; NAL: Nalidixic acid; FOS: Fosfomycin
Author Declaration:
Financial or Other Competing Interests: None
Was Ethics Committee Approval obtained for this study? Yes
Was informed consent obtained from the subjects involved in the study? Yes
For any images presented appropriate consent has been obtained from the subjects. NA
Plagiarism Checking Methods: [Jain H et al.]
Plagiarism X-checker: Jan 05, 2021
Manual Googling: Mar 16, 2021
iThenticate Software: Apr 19, 2021 (22%)
[1]. Hardin G, The Tragedy of the Commons [Internet]. Vol. 162, ScienceAmerican Association for the Advancement of Science[cited 2020 May 6]. Pp. 1243-48. Available from: https://www.jstor.org/stable/172474510.1126/science.162.3859.1243 [Google Scholar] [CrossRef]
[2]. Biggest Threats and Data|Antibiotic/Antimicrobial Resistance|CDC [Internet]. [cited 2020 May 7]. Available from: https://www.cdc.gov/drugresistance/biggest-threats.html [Google Scholar]
[3]. New report calls for urgent action to avert antimicrobial resistance crisis [Internet]. [cited 2020 May 7]. Available from: https://www.who.int/news-room/detail/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis [Google Scholar]
[4]. Ramirez MS, Traglia GM, Lin DL, Tran T, Tolmasky ME, Plasmid-mediated antibiotic resistance and virulence in gram-negatives: The klebsiella pneumoniae paradigmMicrobiol Spectr 2014 2(5):01-15.10.1128/microbiolspec.PLAS-0016-20134335354 [Google Scholar] [CrossRef] [PubMed]
[5]. Langella P, Zagorec M, Ehrlich SD, Morel-deville F, Intergeneric and intrageneric conjugal transfer of plasmids PAM p 1, pIL205 and pIPSO in Lmtobacillus sakeFEMS Microbiology letters 1996 :13910.1111/j.1574-6968.1996.tb08178.x [Google Scholar] [CrossRef]
[6]. Moore ERB, Hoffmann A, Thimm T, Dro M, Munch JC, Tebbe CC, Intergeneric Transfer of Conjugative and Mobilizable Plasmids Harbored by Escherichia coli in the Gut of the Soil Microarthropod Folsomia candida (Collembola)Appl Environ Microbiol 1998 64(7):2652-59.10.1128/AEM.64.7.2652-2659.19989647844 [Google Scholar] [CrossRef] [PubMed]
[7]. Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV, Evidence for transfer of CMY-2 AmpC- Lactamase plasmids between Escherichia coli and salmonella isolates from food animals and humansAntimicrob Agents Chemother 2001 45(10):2716-22.10.1128/AAC.45.10.2716-2722.200111557460 [Google Scholar] [CrossRef] [PubMed]
[8]. Datta N, Hughes VM, Plasmids of the same Inc groups in enterobacteria before and after the medical use of antibioticsNature 1983 306(5943):616-17.10.1038/306616a06316165 [Google Scholar] [CrossRef] [PubMed]
[9]. Couturier M, Bex F, Bergquist PL, Maas WK, Identification and classification of bacterial plasmidsMicrobiol Rev 1988 52(3):375-95.10.1128/mr.52.3.375-395.19883054468 [Google Scholar] [CrossRef] [PubMed]
[10]. Carattoli A, Plasmids in Gram negatives: Molecular typing of resistance plasmidsInt J Med Microbiol 2011 301(8):654-58.10.1016/j.ijmm.2011.09.00321992746 [Google Scholar] [CrossRef] [PubMed]
[11]. Available from: https://www.researchgate.net/profile/Yiyun_Liu6/publication/303234561_Emergence_of_plasmid-mediated_colistin_resistance_mechanism_MCR-1_in_animals_and_human_beings_in_China_a_microbiological_and_molecular_biological_study/links/5d20470692851cf44068f8f9/Em. [cited 2020 May 9] [Google Scholar]
[12]. Huang Y, Yu X, Xie M, Wang X, Liao K, Xue W, Widespread dissemination of carbapenem-resistant escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in ChinaAntimicrob Agents Chemother 2016 60(7):4364-68.10.1128/AAC.00859-1627114282 [Google Scholar] [CrossRef] [PubMed]
[13]. Wang M, Sahm DF, Jacoby GA, Hooper DC, Emerging plasmid-mediated quinolone resistance associated with the qnr Gene in Klebsiella pneumoniae clinical isolates in the United StatesAntimicrob Agents Chemother 2004 48(4):1295-99.10.1128/AAC.48.4.1295-1299.200415047532 [Google Scholar] [CrossRef] [PubMed]
[14]. Alba P, Leekitcharoenphon P, Franco A, Feltrin F, Ianzano A, Caprioli A, Molecular epidemiology of mcr-encoded colistin resistance in Enterobacteriaceae from food-producing animals in Italy revealed through the EU harmonized antimicrobial resistance monitoringFront Microbiol 2018 12:910.3389/fmicb.2018.0121729951045 [Google Scholar] [CrossRef] [PubMed]
[15]. Magiorakos AP, Srinivasan A, Careyetal RB, Carmeli Y, Falagas ME, Giske CG, Multidrugresistant, extensively drug-resistant and pandrug- resistant bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired ResistanceMicrobiology 2011 18(3):268-81.10.1111/j.1469-0691.2011.03570.x21793988 [Google Scholar] [CrossRef] [PubMed]
[16]. Singh SP, Manish R, Sourav S, Fosfomycin: An effective treatment modality for multidrug resisant enterobacteriaceae urinary tract infectionInt J of Sci Res 2018 (3):442-44. [Google Scholar]
[17]. Baral P, Neupane S, Marasini B, Ghimire K, Lekhak B, Shrestha B, High prevalence of multidrug resistance in bacterial uropathogens from Kathmandu, NepalBMC Res Note 2012 5(1):3810.1186/1756-0500-5-3822260454 [Google Scholar] [CrossRef] [PubMed]
[18]. Ahmed S, Jakribettu R, Koyakutty S, Urinary tract infections-an overview on the prevalence and the anti-biogram of gram negative uropathogens in a tertiary care center in North Kerala, IndiaJ Clin Diagn Res 2012 6(7):1192-95. [Google Scholar]
[19]. Aydin A, Ahmed K, Zaman I, Khan MS, Dasgupta P, Recurrent urinary tract infections in womenInt Urogynecol J 2015 26(6):795-804.10.1007/s00192-014-2569-525410372 [Google Scholar] [CrossRef] [PubMed]
[20]. Dolinsky AL, M100 Performance Standards for Antimicrobial Susceptibility TestingJ of Services Marketing 2017 :27-39. [Google Scholar]
[21]. Carattoli A, Elena VR, MINI review resistance plasmid families in EnterobacteriaceaeAntimicrob Agents Chemother 2009 53(6):2227-38.10.1128/AAC.01707-0819307361 [Google Scholar] [CrossRef] [PubMed]
[22]. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ, Identification of plasmids by PCR-based replicon typingJ Microbiol Methods 2005 6(3):219-28.10.1016/j.mimet.2005.03.01815935499 [Google Scholar] [CrossRef] [PubMed]
[23]. Huang XZ, Frye JG, Chahine MA, Glenn LM, Ake JA, Su W, Characteristics of Plasmids in Multi-Drug-Resistant Enterobacteriaceae isolated during prospective surveillance of a newly opened hospital in Iraq. Morgan D, editorPLoS One 2012 7(7):e4036010.1371/journal.pone.004036022808141 [Google Scholar] [CrossRef] [PubMed]
[24]. Johnson TJ, Wannemuehler YM, Sara J, Logue CM, White DG, Nolan LK, Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates plasmid replicon typing of commensal and pathogenic Escherichia coli isolatesAppl Environ Microbiol 2007 73(6):1976-83.10.1128/AEM.02171-0617277222 [Google Scholar] [CrossRef] [PubMed]
[25]. Boot M, Raadsen S, Savelkoul PH, Vandenbroucke-Grauls C, Rapid plasmid replicon typing by real time PCR melting curve analysisBMC Microbiol 2013 13(1):8310.1186/1471-2180-13-8323586392 [Google Scholar] [CrossRef] [PubMed]
[26]. Ranjan M, Karade S, Rahi P, Singh SP, Sen S, Urosepsis due to Multi Drug Resistant Myroides odoratimimus: A case reportInt J Curr Microbiol App Sci 2017 6(8):1930-35.10.20546/ijcmas.2017.608.228 [Google Scholar] [CrossRef]
[27]. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ, Urinary tract infections: Epidemiology, mechanisms of infection and treatment optionsNat Rev Microbiol 2015 13(5):269-84.10.1038/nrmicro343225853778 [Google Scholar] [CrossRef] [PubMed]
[28]. Tandogdu Z, Wagenlehner FME, Global epidemiology of urinary tract infectionsCurr Opin Infect Dis 2016 29(1):73-79.10.1097/QCO.000000000000022826694621 [Google Scholar] [CrossRef] [PubMed]
[29]. Lu PL, Liu YC, Toh HS, Lee YL, Liu YM, Ho CM, Epidemiology and antimicrobial susceptibility profiles of Gram-negative bacteria causing urinary tract infections in the Asia-Pacific region: 2009-2010 results from the Study for Monitoring Antimicrobial Resistance Trends (SMART)Int J Antimicrob Agents 2012 40(Suppl. 1):S37-43.10.1016/S0924-8579(12)70008-0 [Google Scholar] [CrossRef]
[30]. Zowawi HM, Harris PN, Roberts MJ, Tambyah P, Schembri M, Pezzani MD, The emerging threat of multidrug-resistant Gram-negative bacteria in urologyNat Rev Urol 2015 12(10):570-84.10.1038/nrurol.2015.19926334085 [Google Scholar] [CrossRef] [PubMed]
[31]. Levy SB, Marshall B, Antibacterial resistance worldwide: Causes, challenges and responsesReview 2004 10(12):122-29.10.1038/nm114515577930 [Google Scholar] [CrossRef] [PubMed]
[32]. Manikandan S, Ganesapandian S, Singh M, Kumaraguru K, Emerging of multidrug resistance human pathogens from urinary tract infectionsCurr Res Bacteriol 2011 4(1):09-15.10.3923/crb.2011.9.15 [Google Scholar] [CrossRef]
[33]. Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious DiseasesClin Infect Dis 2011 52(5):103-20.10.1093/cid/ciq25721292654 [Google Scholar] [CrossRef] [PubMed]
[34]. Pulcini C, Mohrs S, Beovic B, Gyssens I, Theuretzbacher U, Cars O, Forgotten antibiotics: A follow-up inventory study in Europe, the USA, Canada and AustraliaInt J Antimicrob Agents 2017 49(1):98-101.10.1016/j.ijantimicag.2016.09.02927887966 [Google Scholar] [CrossRef] [PubMed]
[35]. Carattoli A, Miriagou V, Bertini A, Loli A, Colinon C, Villa L, replicon typing of plasmids encoding resistance to newer β-LactamsEmerg Infect Dis 2006 12(7):1145-48.10.3201/eid1207.05155516836838 [Google Scholar] [CrossRef] [PubMed]