Utility of Nucleic Acid Extraction Free COVID-19 Real Time PCR Protocol in Resource Limited Setting: A Pilot Study
Santosh Karade1, Pratik Thosani2, Prashant Patil3, Kavita Bala Anand4, Sourav Sen5, Rajiv Mohan Gupta6
1 Associate Professor, Department of Microbiology, Armed Forces Medical College, Pune, Maharashtra, India.
2 Resident, Department of Microbiology, Armed Forces Medical College, Pune, Maharashtra, India.
3 Resident, Department of Microbiology, Armed Forces Medical College, Pune, Maharashtra, India.
4 Associate Professor, Department of Microbiology, Armed Forces Medical College, Pune, Maharashtra, India.
5 Professor and Head, Department of Microbiology, Armed Forces Medical College, Pune, Maharashtra, India.
6 Professor and Dean, Department of Microbiology, Armed Forces Medical College, Pune, Maharashtra, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Sourav Sen, Professor and Head, Department of Microbiology, AFMC, Pune, Maharashtra, India.
E-mail: sensourav@hotmail.com
Introduction
Coronavirus Disease-19 (COVID-19), a respiratory infection, caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), was first identified in Wuhan, Hubei province, China in December 2019. Alarming increase in the number of cases has put tremendous pressure on existing health resources. Real Time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR), a molecular diagnostic method, is considered gold standard for diagnosis of SARS-CoV-2 infection. It involves Ribonucleic Acid (RNA) extraction as the preliminary step. Innovations to cut down cost and time involved in SARS-CoV-2 testing are need of hour.
Aim
To assess the feasibility of Nucleic Acid Extraction Free (NEF) protocol for COVID-19 diagnosis in resource limited settings.
Materials and Methods
In this pilot study, a panel of 148 Nasopharyngeal (NP) samples was subjected to the novel NEF RT-PCR protocol and results were compared to gold standard RT-PCR on RNA extracted from NP specimen. The cycle threshold value (Ct value) for each target was tabulated in MS Excel Spreadsheet and data analysis was performed using Statistical Package for Social Sciences (SPSS) software version 15.0.
Results
Out of 148 collected samples, 120 showed amplification of E and RNA polymerase gene (RdRp) targets by RNA extraction-based RT-PCR. Overall sensitivity and specificity observed for NEF protocol was 43.94% and 96.42%, respectively.
Conclusion
Further refinement in the protocol would be required to improve the sensitivity of NEF protocol and widespread use in laboratories.
Coronavirus, Naso-pharyngeal swab, Pandemic, Polymerase chain reaction, Respiratory infection
Introduction
Coronavirus Disease-19 (COVID-19) is a respiratory infection caused by SARS-CoV-2 was first identified in Wuhan, Hubei province, China in December 2019 [1]. Globally, the unprecedented increase in the number of COVID-19 cases to over 104 million till January 2020 has resulted in tremendous pressure on existing health resources [2]. Despite several precautionary measures and forewarning, the number of COVID-19 cases is increasing at an alarming rate. Till January 2020, the COVID-19 cases in India increased to 10.8 million, the third largest globally [2]. To combat this pandemic, the country must face challenges of creating mass quarantine facility, COVID-19 dedicated hospitals and most importantly expanding diagnostic capabilities. RT-PCR based diagnosis remains the cornerstone for early diagnosis and management of COVID-19 cases. The RT-PCR methodology typically consist of RNA extraction from respiratory samples, preparation of master-mix, additional of template, followed by real time PCR and analysis of results [3].
The country rose to the challenge by establishing network of viral research and diagnostic laboratories and currently over 530 laboratories are operationalised [4]. The numbers are evolving with increasing participation of government and private laboratories. The column-based RNA extraction costs approximately Rupees (INR) 400/sample in India and remains a major time consuming step. Automated RNA extraction system can handle 24 to 96 samples in an hour; however, the cost remains a limiting factor. Recently, Grant PR et al., and Smyrlaki I et al., described RNA extraction free protocol for real-time PCR [5,6]. If proven effective, NEF protocol would save time and resources. Thus, the aim of the present study was to assess the feasibility of Nucleic Acid Extraction Free (NEF) protocol for COVID-19 diagnosis in resource limited settings.
Materials and Methods
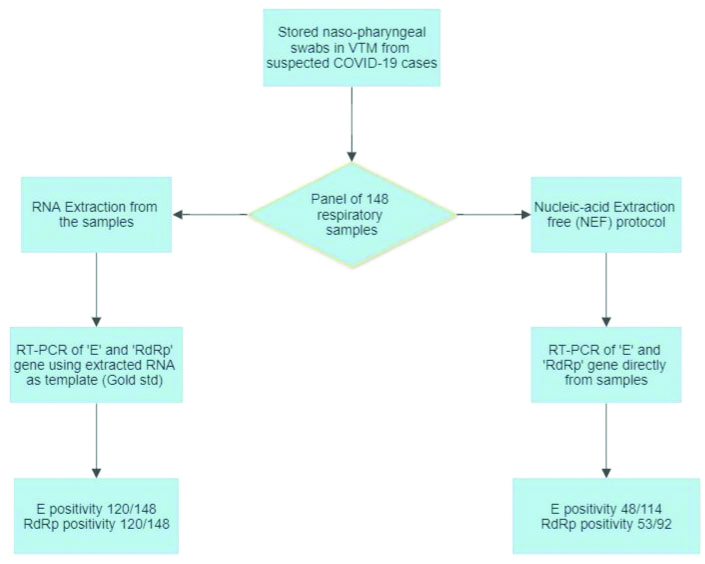
This cross-sectional pilot study was carried out at Indian Council of Medical Research (ICMR) approved COVID-19 diagnostic laboratory of western India. The sample consisted of Nasopharyngeal (NP) swab in 3 mL of Viral Transport Medium (VTM) collected previously from suspected COVID-19 cases and stored at -70°C. Being a pilot study to validate a previously described protocol by Grant PR et al., a sample size of approximately 150 was considered [5]. However, 2 samples failed RNA extraction and thus a final panel of randomly assigned 148 samples were selected. Waiver of ethical clearance for the study was obtained as the study was performed on stored samples without inclusion of any patient level data.
RNA extraction from NP sample was performed using QIAamp viral RNA mini kit (Qiagen) as per manufacturers instruction with final elution volume of 30 μL. RT-PCR for qualitative detection of E Sarbeco (E) gene and RNA dependent RNA polymerase gene (RdRp) was carried out using InvitrogenTM SuperScriptTM III Platinum One-Step qRT PCR Kit. Primer-probe targeting Ribonuclease P (RNase P), a ribozyme found in human cell was utilised as internal control for each PCR. Briefly, 20 μL master mix per reaction containing 5.5 μL of nuclease free water (Qiagen), 0.5 μL Platinum Taq Deoxyribonucleic Acid (DNA) Polymerase. 12.5 μL 2X ready reaction mix (InvitrogenTM), and 1.5 μL of primer probe mix was prepared. To this 20 μL of master-mix, a 5 μL of template RNA extracted from respective sample was added. The primers and probes as shown in [Table/Fig-1] and suggested by World Health Organisation (WHO) were utilised in the study [3,7]. The real-time RT-PCR protocol for amplification of SARS-CoV-2 targets is shown in [Table/Fig-2] [8].
Primer and Probes used for real-time PCR based detection of SARS-CoV-2 in respiratory sample [3,7].
| Gene | Primer and probe | Sequence (5’-3’) |
|---|
| E Gene | E_Serbaco_F1 | ACAGGTACGTTAATAGTTAATAGCGT |
| E_Serbaco_R2 | ATATTGCAGCAGTACGCACACA |
| E_Serbaco_P1 | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ |
| RNase PGene | RNase P Forward | AGATTTGGACCTGCGAGCG |
| RNase P Reverse | GAGCGGCTGTCTCCACAAGT |
| RNase P Probe | FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ |
| RdRp | RdRp Forward | GGTAACTGGTATGATTTCG |
| RdRp Reverse | CTGGTCAAGGTTAATATAGG |
| RdRp Probe | FAM-TCATACAAACCACGCCAGG-BHQ |
Real time PCR cycling conditions for detection of SARS-CoV-2 [8].
| Steps | Process | Temperature | Duration |
|---|
| 1 | Reverse transcription | 55°C | 30 min |
| 2 | Taq inhibitor inactivation | 95°C | 3 min |
| 3 | PCR amplification (45 cycles) | 95°C for 15 sec58°C for 30 sec | Data collection at 58°C |
For Nucleic Acid Extraction Free (NEF) protocol, under appropriate biosafety precautions 5 μL of VTM based sample was directly added to 20 μL of RT-PCR master-mix, as described above, in a Class 2b biosafety cabinet. Similar RT-PCR kit and PCR cycling conditions were used for both the methods.
All 148 samples were subjected to RT-PCR for SARS-CoV-2 by both the methods as shown in flowchart [Table/Fig-3]. The cycle threshold value (Ct value) obtained for E and RdRp gene target [Table/Fig-4] from extracted RNA based RT-PCR and NEF PCR protocol were tabulated in Microsoft Excel spreadsheet and compared.
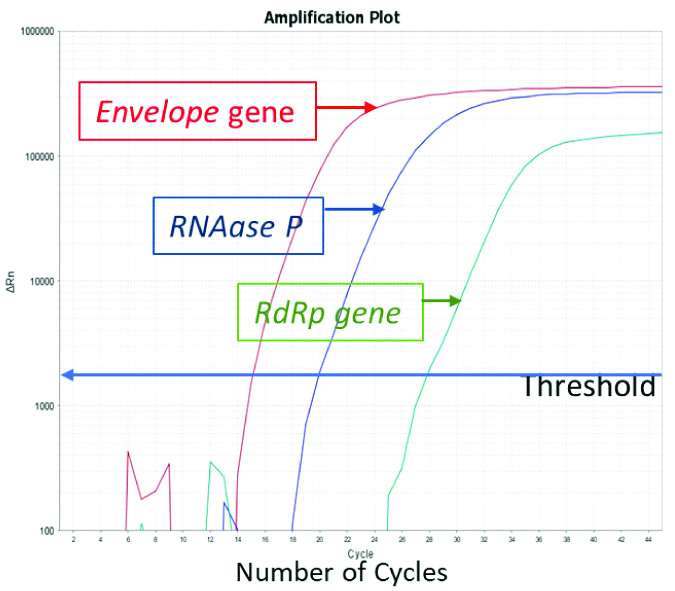
Real Time PCR amplification plot for E gene, RdRp gene and RNase P gene.
Statistical Analysis
Data analysis was performed using SPSS software version 15.0. The RNA extraction-based SARS-CoV-2 RT-PCR method was considered gold standard in this study.
Results
From the panel of 148 previously collected samples, a total of 120 showed amplification of E and RdRp targets by RNA extraction-based RT-PCR. The median (IQR) Ct value obtained for E and RdRp targets for 120 samples was 27 (22-30) and 27.5 (22.2-31), respectively. The internal Control RNase P was amplified in all samples, median (IQR) Ct value; 27 (25.1-29).
In the NEF, RT-PCR protocol, based on successful amplification of internal control RNase P, results of 114 E gene and 92 RdRp gene were considered valid. A total of 48 out of 114 (42.10%) SARS-CoV-2 positive respiratory samples subjected to NEF RT-PCR protocol indicated concordant results with E gene RT-PCR from extracted RNA whereas, 53 of 92 (57.60%) sample indicated concordance for RdRp gene. The median (IQR) Ct value obtained for E and RdRp targets were 31.4 (27.5-35.5) and 33.4 (28.8-36.1), respectively. The median Ct value for E and RdRp target by NEF protocol was higher by 4.4 and 5.9 cycles, respectively as compared to the Gold standard method. For 28 SARS-CoV-2 negative samples, the results of RdRp gene RT PCR matched completely by both the methods.
Whereas, for E gene 27 out of 28 samples showed concordant results. The RNA extraction-based SARS-CoV-2 RT-PCR method was considered gold standard in this study which has an estimated sensitivity and specificity of 70% and 95%, respectively [9]. Of the 92 samples that showed amplification of E and RdRp gene target by Gold Standard, complete concordance was seen in 29 isolates wherein, both E and RdRp gene targets were amplified. A total of 37 samples showed complete discordance and 26 showed partial discordance. The overall sensitivity and specificity of extraction free protocol was 43.94% and 96.42% respectively, when successful detection of both E and RdRp target was considered [Table/Fig-5].
Performance of Nucleic-acid Extraction Free (NEF) PCR protocol.
| PCR target | Results (n) | Test performance |
|---|
| TP | TN | FP | FN | Sensitivity | Specificity |
|---|
| E gene | 48 | 27 | 1 | 66 | 42.10 | 96.42 |
| RdRp gene | 53 | 28 | 0 | 39 | 57.6 | 100 |
| Both E and RdRp | 29 | 27 | 1 | 37 | 43.94 | 96.42 |
TP: True positive; TN: True negative; FP: False positive; FN: False negative. Denominator for E gene target: n=142 (114 positives and 28 negative). Denominator for RdRp gene target: n=120 (92 positives and 28 negative). Denominator for both E and RdRp gene target: n=94
Discussion
Real time PCR based tests are gold standard for diagnosis of SARS-CoV-2 infection responsible for COVID-19 [10]. Considering highly infectious nature and droplet mode of transmission of the pathogen, specific biosafety precautions need to be observed for conducting the diagnostic test. The diagnostic test requires specialised infrastructure, sophisticated equipment, trained manpower and set of standardised protocols. The unprecedented increase in number of cases despite lockdown measures has placed tremendous pressure on health care establishments. The standard test takes an average of 5-6 hours from receipt of batch of samples to getting results. Scaling up of SARS-CoV-2 diagnostic laboratories and decrease in turn-around-time for test results is important for early diagnosis, instituting isolation measures, contact tracing and ultimately breaking the chain of transmission [11].
RNA extraction is pre-requisite for RT-PCR and it takes about 45-60 minutes by silica column based method. A nucleic acid extraction sparing protocol would save time and financial burden in resource limited setting. Bacterial colony PCR, wherein bacterial colony is directly added to PCR master mix is a known practice [12]. However, extraction free PCR for viral studies is not met with much success. In this study assessment for feasibility of extraction free protocol in COVID-19 testing as described previously Grant PR et al., and Smyrlaki I et al., was assessed [5,6]. A 5 μL of respiratory sample in VTM was utilised directly as template for COVID-19 RT-PCR and the results obtained by PCR using 5 μL of RNA as template were compared. The lower Ct value obtained for extraction free protocol can be explained by effect of dilution of NP swab in 3 mL of VTM.
The overall sensitivity and specificity of extraction free protocol was 43.94% and 96.42%, respectively. Grant PR et al, achieved a sensitivity and specificity of 98% and 100% respectively with protocol using 2 μL sample addition to master-mix [5]. As 25 μL PCR reaction containing 5 μL template and 20 μL master mix is standard protocol across most COVID-19 testing laboratories, 5 μL sample addition protocol was explored. In this study, the sensitivity was lower may be due to difference in prime-probe and the PCR kit utilised. Also, in this study refrigerated stored clinical samples at -70°C were used. The heating protocol to neutralise the virus as recommended Smyrlaki I et al., could not be explored as heating resulted in jellification of sample in viral transport media [6]. Also, the procedure of direct heating of respiratory samples in a thermocycler would require additional biosafety precautions [13]. The study results could not meet desired sensitivity for incorporation into SARS-CoV-2 mass screening program. However, the specificity of the protocol was above 96%. In addition, NEF protocol reduced the turn-around time of SARS-CoV-2 RT-PCR by an hour.
Limitation(s)
The study results should be interpreted considering the limitations of sample size. The time of sampling of patient, clinical symptoms and the SARS-CoV-2 viral load in the sample directly affects the results of NEF protocol. The study was performed on stored samples and follow-up patient data was not available to assess the outcome.
Conclusion(s)
With increase in positivity rate of SARS-CoV-2 infection in current pandemic, the sensitivity of the test needs to be worked upon. Thus, this extraction free protocol needs to be cautiously evaluated by each laboratory. Also, the biosafety issues associated with directly handling infective samples needs to be approached carefully. Till date, World Health Organisation or Indian Council of Medical Research recommends only the use of RNA extraction method for Real time PCR for COVID-19 testing. Further studies on clinical samples would be necessary to validate NEF protocol as a cost-effective measure in resource limited settings.
TP: True positive; TN: True negative; FP: False positive; FN: False negative. Denominator for E gene target: n=142 (114 positives and 28 negative). Denominator for RdRp gene target: n=120 (92 positives and 28 negative). Denominator for both E and RdRp gene target: n=94
Author Declaration:
Financial or Other Competing Interests: None
Was Ethics Committee Approval obtained for this study? No
Was informed consent obtained from the subjects involved in the study? No
For any images presented appropriate consent has been obtained from the subjects. NA
Plagiarism Checking Methods: [Jain H et al.]
Plagiarism X-checker: Jan 05, 2021
Manual Googling: Jan 23, 2021
iThenticate Software: Apr 04, 2021 (12%)
[1]. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, A Novel Coronavirus from patients with pneumonia in China, 2019N Engl J Med 2020 382:727-33.10.1056/NEJMoa200101731978945 [Google Scholar] [CrossRef] [PubMed]
[2]. World Health Organization, Geneva. Coronavirus disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. Source [Internet]. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ [Google Scholar]
[3]. WHO Protocol: Real-time RT-PCR assays for the detection of SARS-CoV-2. Institut Pasteur, Paris. Available from: https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf [Google Scholar]
[4]. Gupta N, Potdar V, Praharaj I, Giri S, Sapkal G, Yadav P, Laboratory preparedness for SARS-CoV-2 testing in India: Harnessing a network of Virus Research & Diagnostic LaboratoriesIndian J Med Res 2020 151:216-25.10.4103/ijmr.IJMR_594_2032242875 [Google Scholar] [CrossRef] [PubMed]
[5]. Grant PR, Turner MA, Shin GY, Nastouli E, Levett LJ, Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemicbioRxiv 2020 19:2020.04.06.02831610.1101/2020.04.06.028316 [Google Scholar] [CrossRef]
[6]. Smyrlaki I, Ekman M, Vondracek M, Papanicoloau N, Lentini A, Aarum J, Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-qPCRmedRxiv 2020 :2020.04.17.2006734810.1101/2020.04.17.2006734832968075 [Google Scholar] [CrossRef] [PubMed]
[7]. ICMR-SOP_for_Confirmatory_Assay_for_2019_nCoV.pdf. Available from: https://www.icmr.gov.in/pdf/covid/labs/2_SOP_for_Confirmatory_Assay_for_2019_nCoV.pdf [Google Scholar]
[8]. ICMR-SOP_for_First_Line_Screening_Assay_for_2019_nCoV.pdf. Available from: https://www.icmr.gov.in/pdf/covid/labs/1_SOP_for_First_Line_Screening_Assay_for_2019_nCoV.pdf [Google Scholar]
[9]. Watson J, Whiting PF, Brush JE, Interpreting a covid-19 test resultBMJ 2020 :m1808Available at: https://doi.org/10.1136/bmj.m180810.1136/bmj.m180832398230 [Google Scholar] [CrossRef] [PubMed]
[10]. World Health OrganizationLaboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human casesWHO-Interim guidance 2020 2019(January):01-07. [Google Scholar]
[11]. Wee SK, Sivalingam SP, Yap EPH, Rapid direct nucleic acid amplification test without rna extraction for SARS-CoV-2 using a portable PCR thermocyclerGenes 2020 11(6):66410.3390/genes1106066432570810 [Google Scholar] [CrossRef] [PubMed]
[12]. Woodman ME, Savage CR, Arnold WK, Stevenson B, Direct PCR of intact bacteria (colony PCR)Current Protocols in Microbiology 2016 2016(August):A.3D.1-A.3D.7.10.1002/cpmc.1427517337 [Google Scholar] [CrossRef] [PubMed]
[13]. WHO Headquarters (HQ), WHO Worldwide. Protocol for assessment of potential risk factors for coronavirus disease 2019 (COVID-19) among health workers in a health care setting. 25 Jan 2020. Source: Internet. Available at: https://www.who.int/publications/i/item/protocol-for-assessment-of-potential-risk-factors-for-2019-novel-coronavirus-(2019-ncov)-infection-among-health-care-workers-in-a-health-care-setting [Google Scholar]