Introduction
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2) virus, a causative agent of COVID-19 has led to universal pandemic. During this pandemic there has been an acute shortage of good quality Viral Transport Medium (VTM) because of increase in number of infected people worldwide. It is also difficult to maintain the transport and storing conditions in line with the guidelines in pandemics.
Aim
To assess the feasibility of Oropharyngeal Swab (OP)/Nasal swabs in 0.9% normal saline in place of VTM and to analyse the effect of temperature on nucleic acid detection by real time Reverse Transcription Polymerase Chain Reaction (rRT PCR) on saline samples stored at 4°C, ambient and at higher temperature (37°C).
Materials and Methods
The present study was an observational analytical study which included 94 positive and 5 negative samples. Patients’ nasal or OP samples were collected as dry swabs and in VTM. Normal saline was added once the samples were received in the laboratory. Polymerase Chain Reaction (PCR) was done with saline and VTM samples both on day 1. Samples were aliquotted in 3 sets and one set was kept at 4°-8° C and other two at 25°C and 37°C, respectively. All positive samples were further tested on day 3, day 4 and day 6. Results were analysed and compared.
Results
Samples in normal saline showed very good sensitivity at all temperatures (4°-8°C, 25°C and 37°C) till day 6. Both the swab samples (in saline and in VTM) showed nearly 100% agreement in rRT-PCR results. Cycle threshold (Ct) value variation was also ±2.
Conclusion
Looking into the cost and logistics issues especially during pandemics, saline is a good and cheaper alternative to VTM and with its use, testing capacity can be expanded.
Introduction
The SARS CoV-2 was first detected and described in December 2019 in the Wuhan city of China, as the causative agent of COVID-19 and has led to global spread causing universal pandemic [1]. Infected patients show varied symptoms ranging from absence of symptoms to severe pneumonia requiring Intensive Care Units (ICUs) and fatal outcomes. Early diagnosis would aid in timely treatment of the patient and isolating such patients can help in decreasing further spread [2].
The standard diagnostic line of testing the presence of SARS CoV-2 in patients is based on collecting pharyngeal swab samples in a transport media from which viral Ribonucleic Acid (RNA) is extracted and Real time PCR is done [1]. It is considered as the gold standard test for this disease [3].
Samples that are considered to be important for laboratory diagnosis of COVID-19 disease are OP, Nasopharyngeal swabs, Nasal swabs, sputum, lower respiratory tract aspirates, Broncho Alveolar Lavage (BAL), Nasopharyngeal wash/aspirate or Nasal aspirate [4]. Out of these Oropharyngeal and Nasopharyngeal swabs are among the commonly used ones and are collected in commercial VTM. VTM consists of Hanks Balanced salt solution, foetal bovine serum, antibiotics and antifungals [5] and is used for transportation of viruses while maintaining the viability.
Following criteria should be fulfilled by any media which is to be used for specimen collection and transport for RT PCR- detection of SARS CoV-2 [6]:
Compatibility with molecular diagnostics and further genome analysis.
No degradation of nucleic acid.
Can be easily used in field settings, while preserving microbial nucleic acid.
Cheaper and easily available.
Also, World Health Organisation (WHO), Centre for Disease Control and prevention (CDC), European Center for Disease Prevention and Control (ECDC) and other Health authorities have emphasised that the accuracy of RT-PCR results depends upon proper specimen collection and storage [5,7,8]. Proper specimen collection refers to proper transport media and proper way of sample collection taking all precautions and storage infers to maintaining temperature of 4°C to 8°C, until the sample processing is started for Real time PCR.
During 2020, SARS CoV-2 pandemic there has been an acute shortage of good quality VTM supply [9] due to increase in number of infected people in such a short period of time [10] because of movement of people and goods around the world. To meet the increased demand many different reagents have been tried and assessed to meet large scale diagnostics and surveillance testing [9].
In a developing, over populated country like India, with such an outrageous number of samples during this pandemic, apart from availability of VTM which is severely limited, it is difficult to maintain the transport and storing conditions in line with the guidelines [11]. The logistics to arrange and transport the samples to laboratory, sometimes hundreds of miles away from the laboratory can be cumbersome and resource intensive [12]. All of these factors contribute to delay in diagnostics and rationing of diagnostic testing [13].
For molecular testing, preserved nucleic acid are needed rather than replication competent viruses [12]. But for culture and further testing with the isolates, live and intact viruses are necessary for which we need to collect and transport samples in good quality VTM. Kirkland PD and Frost MJ have shown that many commercial VTMs contain nucleases and similar substances which can seriously compromise the results of diagnostic tests [9]. They may be suitable for virus culture purposes but with nucleic acid based tests, the results can be misleading. So, all the products should be evaluated for ensuring fitness for the purpose.
Apart from this, the effect of temperature on virus’ survivability has quoted [14], but little is known about the impact of temperature on nucleic acid detection by PCR.
Thus, this study was aimed to assess the feasibility of OP/Nasal swabs in 0.9% normal saline after collecting them as dry swabs at the point of collection and performance of RT-PCR on the samples collected in VTM and dry swabs dipped in normal saline. Secondly, to analyse the effect of temperature on nucleic acid detection by RT PCR on saline samples stored at 4°C, ambient as well as at higher temperature (37°C). Latter two temperatures mimicked field conditions in which specimens remain for hours.
Materials and Methods
Sample collection and storage
The present study was an observational analytical study conducted in Department of Microbiology, SMS Medical College Jaipur in December, 2020. The data was analysed from the records and selected 94 positive patients showing a wide range of Ct values and 5 negative patients. Their fresh samples were collected, one in VTM and other as dry swab. Paired identical nasal swab or OP specimens were collected from each patient in VTM (Vitromed healthcare, Biotech Park, Jaipur, Rajasthan) and empty and sterile 15 mL falcon tube, respectively. The patient samples showing Ct value >35 after retesting, were excluded from the study. Patient’s verbal consent was taken at the time of sample collection and rest the study was done on the collected samples, patients’ details were not disclosed.
Once the samples were received in the laboratory, 3.5 mL of sterile normal saline (0.9% W/V manufactured by RUSOMA Lab Pvt., ltd., Indore) was added to the falcon tubes in which dry swabs were collected, after that it was vortexed well. Sample was added to the sample plate for RNA extraction by automated extraction system (Perkins Elmer chemagic 360), as per the manufacturers’ instruction. This whole procedure was carried out in Biosafety Level-2 laboratory. At the same time saline tube samples were aliquotted in three sets of eppendorf tubes (1 ml volume in each) after thorough vortexing. Each set of saline aliquots were kept at 4°C, ambient temperature (23°-26°C) and 37°C (Incubator) respectively. Refrigerator and incubator temperatures were monitored by thermometer regularly. In addition to these samples, authors took five negative samples also, which were stored under same conditions as a control group.
First RT-PCR test of these samples was done on the day sample were received in the laboratory i.e., day One (D1) within 24 hours of collection. After that, all the samples (stored at 4°C, ambient temperature 23°C- 26°C and 37°C) were tested on intervals of 72 hours, 120 hours, and 144 hours). Automated extraction system (Perkins Elmer Chemgic 360) was used for RNA extraction followed by RT-PCR by TruPCR SARS-COV-2 detection kit as per manufacturer’s instructions. Ct values of all the samples’ results were noted and compared.
Extraction
After thorough vortexing, followed by brief centrifugation of the VTM/saline tubes, 300 μL of the sample was transferred to a 96 deep well processing plate to which 4 μL Poly (A) RNA, 10 μL of proteinase K, 300 μL lysis buffer along with 150 μL magnetic beads and 900 μL of RNA binding buffer were already been added. The beads/RNA mixture was washed with washing buffer and elutes were obtained in elution buffer in the automated system (Perkins Elmer Chemagic 360).
Real time PCR (Tru PCR master mix)
The primers used in TruPCR RT PCR kit are designed to target E gene, N/RdRp and Rnase Pgenes. For PCR, 10 uL RNA and 15 μL PCR master mix solution containing 10 uL master mix reagent, 0.35 μL Enzyme mix and 4.65 μL of primer probe mix. Cyclic conditions used as per the manufacturer’s instructions were 50°C for 15 minutes, 95°C for 5 minutes, then 38 repeat cycles of 95°C for 5 seconds, 60°C for 40 seconds and 72°C for 15 seconds, using Biorad CFx platform.
Statistical Analysis
The statistical software-Statistical Package for the Social Science (SPSS) version 20 was used for the analysis of the data and Microsoft word and Excel have been used to generate graphs and tables.
Results
A total of 94 paired SARS CoV-19 samples were collected from COVID-19 positive patients and five from COVID-19 negative people and PCR results were analysed. All positive samples were put in PCR on day 1 (<24 hours of collection), day 3 (72 hrs), day 5 (120 hrs) and day 6 (144 hrs) and negative samples were put in PCR on day 1 and day 6 only.
One positive and one negative internal control were put in each PCR run and their results came within established Quality Control (QC) ranges (data not shown).
All five negative samples gave negative results on PCR with valid graph of Ribonuclease P (RNase P), demonstrating that the swabs and saline were free of any kind of SARS CoV-2 contamination.
Overall, both the swabs (in saline and in VTM) showed almost 100% agreement in positivity. Ct value variation was also not much (only ±2 Ct value variation was noted). This stood correct for all the samples irrespective of viral concentration in the sample (considering samples of different ranges of Ct values) (As depicted in [Table/Fig-1]).
Comparison of RT PCR results (RdRp gene Ct Values) of samples in saline and VTM within 24 hours of collection.
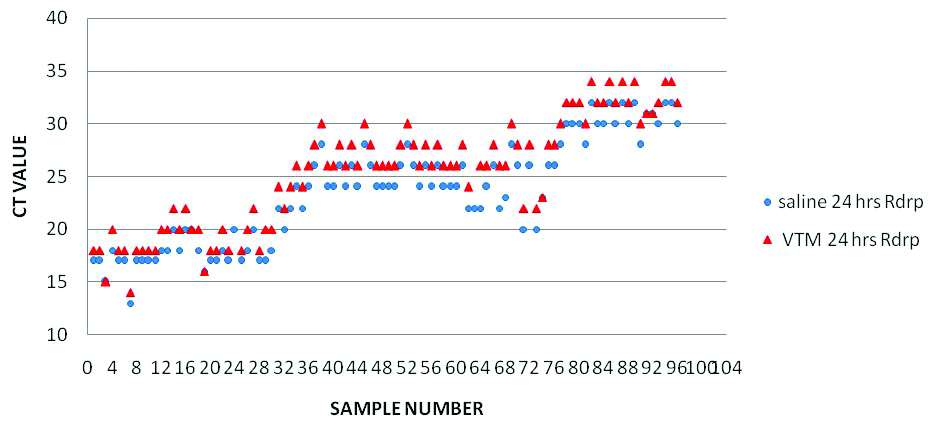
Two samples came positive by rRT-PCR with Saline but invalid with VTM. Two samples gave invalid results on day four at all temperatures.
One sample came negative in 25°C group on day 4 but interestingly came positive on day 5. There was no significant evidence of loss of sensitivity or stability of the sample (>3Ct value increase) except in two samples. There was no sample left with any sample for putting run on Day 7.
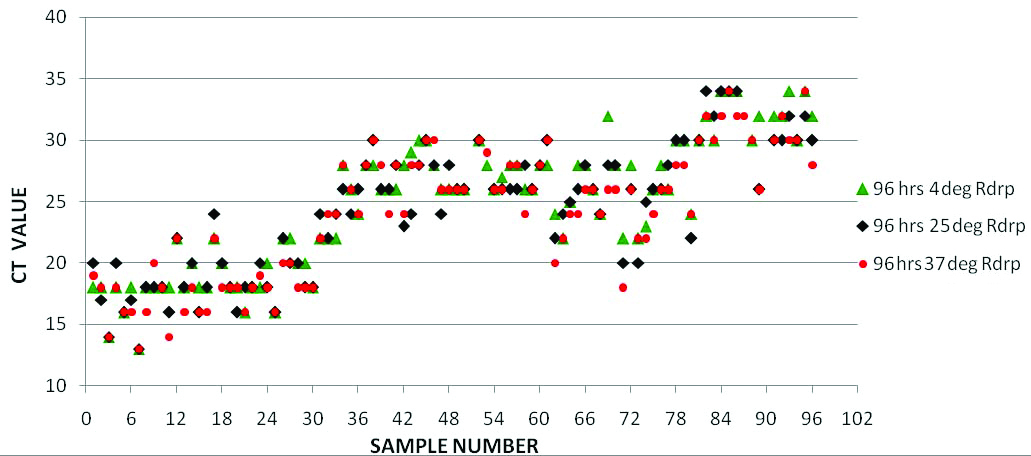
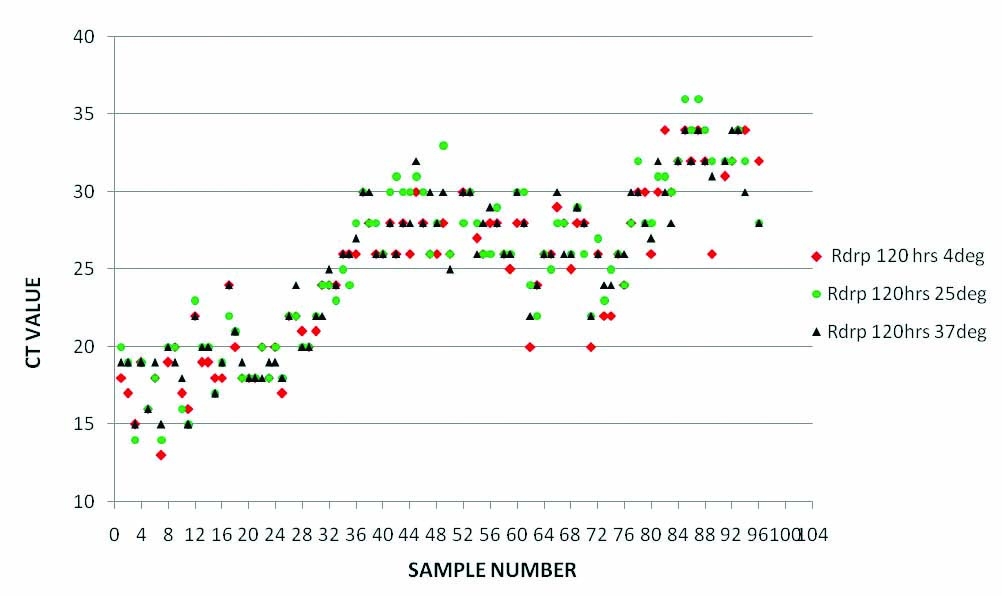
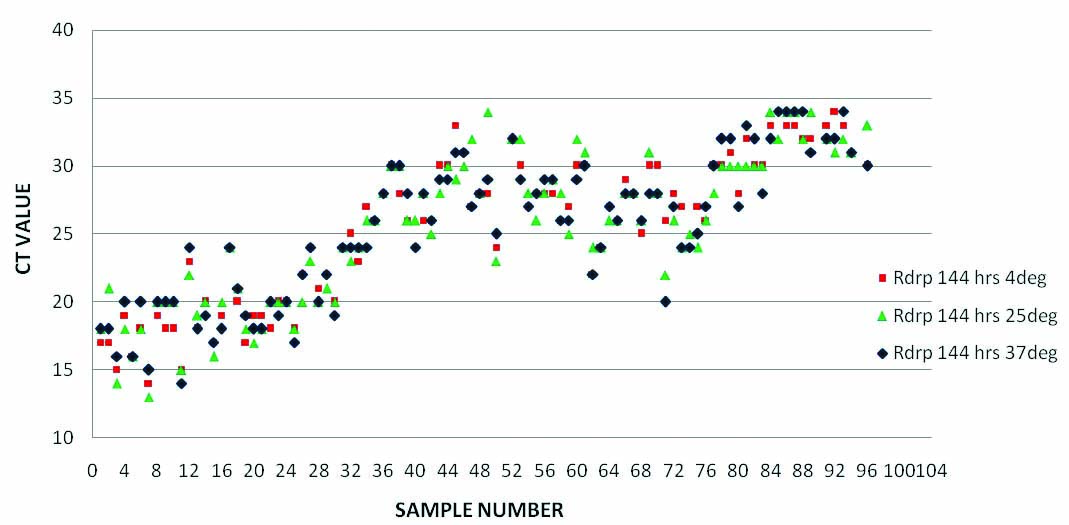
Till day 6, all the saline samples showed very good sensitivity at all the temperatures. To our surprise, results were slightly better (Low Ct) for the samples incubated at 37°C with higher consistency without showing any significant false negativity or variation in Ct values [Table/Fig-2,3 and 4].
Comparison of RT PCR results (RdRp gene Ct Values) of samples in saline at three temperatures (4°C, ambient temperature and 37°C) on fourth day of collection.
Comparison of RT PCR results (RdRp gene Ct Values) of samples in saline at three temperatures (4°C, ambient temperature and 37°C) on fifth day of collection.
Comparison of RT PCR results (RdRp gene Ct Values) of samples in saline at three temperatures (4°C, ambient temperature and 37°C) on sixth day of collection.
Discussion
During epidemics/pandemics, depletion of culture/transport media usually prompts the use of other alternatives because of substantial increase in sample load. It has already been demonstrated that Phosphate Buffered Saline (PBS) can be used in place of VTM for detection of SARS CoV-2 [12-14].
In the present study, plain/dry swabs samples in saline were compared with swabs in VTM for diagnosis of SARS CoV-2 by RT PCR. Saline samples yielded similar results with respect to Ct values as their respective swabs in VTM. This is in concordance with the studies done by Druce J et al., and Rodino KG [15,16]. Both the studies have recommended the use of saline in place of VTM for collection and transport of samples for detection by PCR. This was also recommended by the present study. It is also advocated that the extraction/PCR inhibitors or nucleases might be present in commercial VTM that affect RNA detection during RT PCR [6]. In this study, two of the samples came positive by saline samples but invalid by the corresponding VTM samples. Saline has no such inhibitors, so it can be considered as a better alternative apart from being a cheaper option.
It is recommended by CDC, WHO and ECDC that the samples should be stored at 2°C-8°C for upto 3-5 days and -70°C if more than 5 days’ storage is required [5,7,8]. Maintaining such temperature during shipment and transportation, especially in pandemics/epidemics is cumbersome and involves many cost and logistic issues. Many laboratories tend to reject the samples, if these guidelines are not followed. In this study, no significant effect of ambient/37°C temperatures on SARS CoV-2 RNA was noted for upto 6 days. This is in concordance with studies done by Agaoglu NB and Druce J et al., [11,15]. Former has shown that OP/NP samples in VTM kept at ambient temperature remain positive in SARS CoV-2 PCR test for 5 days and latter has reported that amplifiable results are obtained even if samples are kept at 25°C and 37°C, though it has also been reported in the study that after 3 days at 37°C, there was gradual loss of nucleic acid integrity in four viruses (Influenza A, Herpes Simplex Virus-2 (HSV-2), enterovirus and adenovirus 7), but in this study observe any such loss at 37°C for SARS CoV-2 was not observed. This confers to more resistant/defiant nature of SARS CoV-2 virus.
Wide range of Ct values were taken in this study but all the samples even those with higher Ct values showed almost same Ct values despite incubating them at 37°C for 6 days. This shows that even at 37°C there was no decay in viral signals even over 6 days period.
Researchers have studied the temperature effect on SARS CoV-2 RNA for upto 25°C [11-13,16], but in tropical areas like Rajasthan, most of the times in a year the temperature remains more than 30°C. This is the reason one set of samples were kept in the incubator (37°C) and to analyse the effect. Taking into consideration the data of our study, authors recommend that even if temperature conditions are not fulfilled, the samples can be analysed and reported.
It is unlikely that virus isolation would match the sensitivity of PCR analysis under the chosen conditions [12]. pH of VTM is around 7-7.4 whereas pH of normal saline is 5.5, and this acidic pH may not be suitable for virus isolation purpose but is acceptable for molecular testing. Le Vay K et al., has quoted that RNA is most stable and its integrity is better preserved at pH 4-5, with significant acid hydrolysis not occurring below pH 2 [17]. In this study also, no untoward effect of low pH on sample positivity was found. So for reference laboratory, saline as a collection and transport media and such temperature conditions might not be suitable. However, for all the diagnostic laboratory saline can be considered as a cheaper and reliable option.
At the point of collection, samples were collected as dry swabs in empty and sterile falcon tubes and were transported at ambient temperature to the laboratory. Saline was added to these tubes later when received in the laboratory. The samples were transported in dry condition without any liquid media in it so chances of leakage or cross contamination were minimal. Routinely, VTM samples are transported in thermocol boxes which increases the volume of waste generated and requires extra efforts for disposal. The present study infers that when dry swabs are used, sample transportation can be done in any cardboard box. The need for immediate transport of clinical specimens to laboratory and maintenance of cold chain is also precluded using dry swabs.
Limitation(s)
Further studies are required to access the reproducibility of SARS-CoV-2 RNA detection in saline samples over in tropical climates with higher ambient temperature and humidity.
Conclusion(s)
Looking into cost and logistics issues especially during pandemics, authors conclude that saline is a good and cheaper alternative to VTM. Collection and transportation of samples collected as dry swabs can be done simply without putting extra efforts in packaging and temperature maintenance. This way, testing capacity can be expanded and more widespread surveillance can be done.
[1]. Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser AM, Glass B, SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-lampViruses 2020 12(8):86310.3390/v1208086332784757 [Google Scholar] [CrossRef] [PubMed]
[2]. Rao SN, Manissero D, Steele VR, Pareja J, A systematic review of the clinical utility of cycle threshold values in the context of COVID-19Infect Dis Ther 2020 9(3):573-86.Epub 2020 Jul 28. Erratum in: Infect Dis Ther. 2020 Aug 1810.1007/s40121-020-00324-3 [Google Scholar] [CrossRef]
[3]. Lippi G, Simundic AM, Plebani M, Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19)Clin Chem Lab Med 2020 58(7):1070-76.10.1515/cclm-2020-028532172228 [Google Scholar] [CrossRef] [PubMed]
[4]. Loeffelholz MJ, Tang YW, Laboratory diagnosis of emerging human coronavirus infections- The state of the artEmerg Microbes Infect 2020 9(1):747-56.10.1080/22221751.2020.174509532196430 [Google Scholar] [CrossRef] [PubMed]
[5]. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19), May-22 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html [Google Scholar]
[6]. Daum LT, Worthy SA, Yim KC, Nogueras M, Schuman RF, Choi YW, A clinical specimen collection and transport medium for molecular diagnostic and genomic applicationsEpidemiol Infect 2011 139(11):1764-73.Epub 2010 Dec 1610.1017/S095026881000238421205332 [Google Scholar] [CrossRef] [PubMed]
[7]. Methodology for estimating point prevalence of SARS-CoV-2 infection by pooled RT-PCR testing, 2020. https://www.ecdc.europa.eu/sites/default/files/documents/Methodology-estimating-point-prevalence%20-SARS-CoV-2-infection-pooled-RT-PCR-testing.pdf [Google Scholar]
[8]. Laboratory testing for coronavirus disease (COVID-19) in suspected human cases, March 19, 2020. https://apps.who.int/iris/bitstream/handle/10665/331501/WHO-COVID-19-laboratory-2020.5-eng.pdf?sequence=1&isAllowed [Google Scholar]
[9]. Kirkland PD, Frost MJ, The impact of viral transport media on PCR assay results for the detection of nucleic acid from SARS-CoV-2Pathology 2020 52(7):811-14.10.1016/j.pathol.2020.09.01333250079 [Google Scholar] [CrossRef] [PubMed]
[10]. Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA, The COVID-19 pandemic: A comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and controlJ Clin Med 2020 9(4):122510.3390/jcm904122532344679 [Google Scholar] [CrossRef] [PubMed]
[11]. Agaoglu NB, Yıldız J, Dogan OA, Alkurt G, Kose B, Demirkol YK, COVID-19 PCR test performance for samples stored at ambient temperaturebioRxiv 2020 Jan 1 10.1101/2020.06.15.153882 [Google Scholar] [CrossRef]
[12]. Perchetti GA, Huang ML, Peddu V, Jerome KR, Greninger AL, Stability of SARS-CoV-2 in PBS for molecular detectionJournal of Clinical Microbiology 2020 58(8):e01094-20.10.1128/JCM.01094-20 [Google Scholar] [CrossRef]
[13]. Radbel J, Jagpal S, Roy J, Brooks A, Tischfield J, Sheldon M, Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport mediumThe Journal of Molecular Diagnostics 2020 22(7):871-75.10.1016/j.jmoldx.2020.04.20932405270 [Google Scholar] [CrossRef] [PubMed]
[14]. Xie J, Zhu Y, Association between ambient temperature and COVID-19 infection in 122 cities from ChinaSci Total Environ 2020 724:138201Epub 2020 Mar 3010.1016/j.scitotenv.2020.13820132408450 [Google Scholar] [CrossRef] [PubMed]
[15]. Druce J, Garcia K, Tran T, Papadakis G, Birch C, Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCRJournal of Clinical Microbiology 2012 50(3):1064-65.10.1128/JCM.06551-1122205810 [Google Scholar] [CrossRef] [PubMed]
[16]. Rodino KG, Espy MJ, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E, Evaluation of saline, phosphate-buffered saline, and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testingJ Clin Microbiol 2020 58(6):e00590-20.10.1128/JCM.00590-20 [Google Scholar] [CrossRef]
[17]. Le Vay K, Salibi E, Song EY, Mutschler H, Cover feature: nucleic acid catalysis under potential prebiotic conditionsChemistry-An Asian Journal 2020 15(2):214-30.10.1002/asia.20190120531714665 [Google Scholar] [CrossRef] [PubMed]