The GBS colonising the genital tract of pregnant women can transmit to its neonate and cause infections in them. In developed countries, interventions such as intrapartum antibiotic prophylaxis following antenatal screening for GBS colonisation have been effective in reducing the incidence of such vertical transmissions and neonatal infections [1]. Meagre resource countries have a high rate of neonatal mortality as no definite interventions are being set up for this neonatal health burden. Infections are one of the major cause of most neonatal deaths in the country even though GBS is not known to be the major cause [2]. It has been suggested that even if the prevalence of neonatal sepsis due to GBS is low in presence of high maternal risk factors for transmission to its neonate, intrapartum prophylaxis may be useful [3]. The US CDC recommends antepartum screening at 35-37 weeks of pregnancy to lower vertical transmission of GBS from mothers to newborns and to manage neonatal case burden [4]. Prevailing reports from India shows a maternal GBS colonisation rates from 1.62% up to 15% [5,6] but there is lack of data from Assam. Assam is one of the nine states in India which carries the burden of having higher infant mortality rate viz., 41/1000 live births in Assam compared to 32/1000 live births than the national average [7]. Authors have previously reported detection of GBS in a case of neonatal meningitis in Dibrugarh hence the role of GBS infections in neonates cannot be ignored [8]. This necessitates the need for antenatal screening for GBS colonisation in pregnant women of this state and broadly from this region. In order to study its epidemiology, serotyping is considered as an important tool. This could also help decide vaccine implementation in the era of antibiotic resistance [9,10]. The present study, therefore, aimed to evaluate the colonisation rate and serotypes of GBS isolated from pregnant women in Dibrugarh, Assam, India.
Materials and Methods
This was a hospital based observational study involving pregnant women attending antenatal clinic for routine check-up at the District Urban Health Centre, Dibrugarh, Assam, India. Vaginal swabs were obtained from consenting pregnant women at 35-37 weeks of gestation. The study also involved their neonates delivered at the hospital and the mothers together with their neonates were followed-up in the postnatal wards for any outcomes of interest. The microbiological procedures were carried out at ICMR-RMRC, Dibrugarh, Assam, India.
The study was approved by the Institutional Ethics Committee, ICMR-RMRC, Dibrugarh vide Approval No. RMRC/Dib./IEC (Human)/ 2013-14/3787) to involve pregnant women and newborns as study subjects. Permission was also taken from the Joint Director of Health Services, Dibrugarh, Assam, India. The participant’s consents were sought and gained by explaining the objectives of the study to them and the benefits therein. Demographic information and other relevant obstetric history of the enrolled participants were obtained by structured questionnaires. The study included the pregnant women at their last trimester who consented to participate. The exclusion criteria were not consenting pregnant women, women undergoing any antibiotic treatment in the last two weeks and women with any vaginal bleeding during the time of counselling.
Sample size calculation: This was a hospital based observational study. As there was no prior hypothesis and lack of earlier data from the study site, therefore the study requires no pre-defined sample size. However, all pregnant women with 35-37 weeks of gestation period attending the outpatient ward of the Urban Health Centre, Dibrugarh were targeted to be included within the study period.
Sample collection and processing: Vaginal swabs were collected during the period of July 2014-June 2016 (two years) from the lower vaginal wall of late trimester pregnant women using sterile cotton swab (HiMedia, Mumbai) by a nurse under the supervision of the attending gynaecologist and without using a speculum. The swab was immediately placed into Stuart’s transport media (HiMedia, Mumbai) and transported to the laboratory at room temperature within 5-6 hours.
Isolation and identification of GBS: The transported swab was inoculated into 1ml selective Todd-Hewitt broth for 18-24 hours at 37°C. Subculturing was done on blood agar with 5 % sheep blood (HiMedia, Mumbai) and incubated for 24 hours at 37°C.
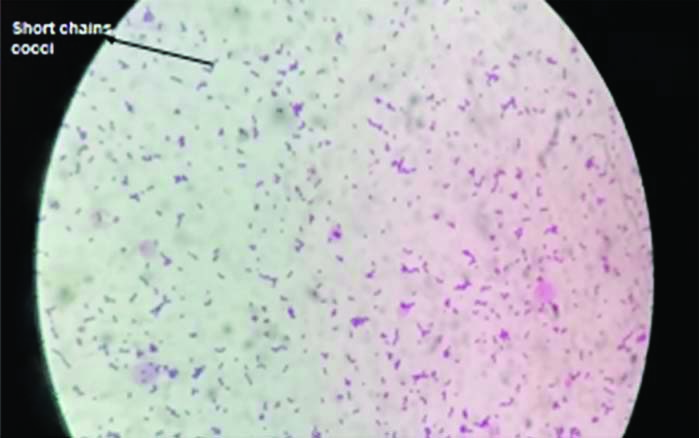
Isolates were grown on blood agar and identified as GBS by following criteria i.e., β-haemolysis on a blood agar plate (few strains were non-haemolytic), gram staining showing gram positive cocci in pairs or short chains [Table/Fig-1], negative reaction with catalase reagent, and Lancefield grouping with group B antisera (Hi-Strep latex agglutination kit).
Microscopic view of gram positive cocci in short chains, viewed under X100 magnification.
The isolates identified through gram staining and biochemical test were then kept for overnight incubation in Brain Heart Infusion (BHI) broth (HiMedia, Mumbai) and preserved at -80°C with addition of 15% glycerol.
Sub-culturing of GBS isolates: Preserved GBS isolates were sub-cultured for further downstreaming processes viz., DNA extraction, preliminary confirmation with scpB gene, serotyping and antibiogram profiling.
Molecular confirmation of GBS isolates by Polymerase Chain Reaction (PCR): Isolates were grown overnight in Brain Heart Infusion (BHI) broth and Deoxyribonucleic Acid (DNA) was extracted isolate wise using Qiagen DNA extraction kit (Qiagen, Germany) following manufacturer’s instructions. PCR targeting the scpB gene that encodes C5a peptidase using previously published primers (3’- ACAACGGAAGGCGCTACTGTT-5’; 3’-ACCTGGTGTTTGACCTGAACT-5’) was carried out and the expected amplified DNA fragment products was of 255 bp [11,12].
Serotyping of GBS isolates: Serotyping of GBS was done by two sets of multiplex PCR using the protocol published elsewhere which targeted the 10 CPS viz., Ia, Ib, II, III, IV, V, VI, VII, VIII and IX [13-15]; primer sets- set 1 for serotypes Ia, Ib, II, III, IV and set 2 for serotypes- V, VI, VII, VIII, IX. All amplifications for serotyping were performed in a Veriti thermal cycler (Applied Biosystems).
Antibiogram profiling: The Kirby Bauer disc diffusion method was used for antimicrobial susceptibility testing. The latest guideline of the CLSI was used for interpretation as resistant, intermediate and sensitive [16]. AST was done on Mueller-Hinton Agar (MHA) with 5% sheep blood. Antibiogram profiling was done for ampicillin (10 μg), penicillin (10 μg), vancomycin (30 μg), levofloxacin (5 μg), erythromycin (15 μg), cefotaxime (30 μg), and clindamycin (2 μg). Streptococcus pneumoniae ATCC 49619 was used as a control strain.
16S rRNA gene amplification from GBS isolates and DNA sequencing: A 1512 base pair partial fragment of 16S rRNA was amplified by PCR employing a pair of universal primers viz., 16S F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16S R (5′-ACGGCTACC TTGTTACGACTT-3′) and using the protocol described earlier by Marin M et al., [17]. Amplified 16S rRNA partial sequences from GBS isolates were gel excised after electrophoresed in 0.7% Agarose (prepared in 1X Tris Acetate EDTA) and purified with QIAEX II Gel Extraction Kit (Qiagen, Germany). Purified PCR products were outsourced to Eurofins Genomics India Private Limited for bi-directional sequencing with Sanger’s dideoxy chain termination chemistry with same set of forward and reverse primer. Out of the 30 serotyped strains, amplification of 16S rRNA sequence was successful for 28 strains and unsuccessful for two strains.
Sequence editing and phylogenetic analysis: Both forward and reverse sequence of each strain were checked and manually edited in BioEdit Program. A total of 28 numbers 16S rRNA gene consensus sequences were created from forward and reverse sequences of each strain with a final size 1440 base pairs. Sequences were checked for presence of chimeras in DECIPHER web tool version 2.17.1 [18]. The 16S rRNA partial sequences of S. agalactiae strains were submitted to GenBank, NCBI under accession numbers MT626729–MT626756.
Phylogenetic relationship among S.agalactiae strains in this study was inferred in Molecular Evolutionary Genetics Analysis (MEGA) 7.0 [19]. For phylogeny, authors retrieved 16S rRNA sequence of E.coli (accession number NR_024570) from National Center for Biotechnology Information (NCBI) and considered E.coli strain as out group. Multiple sequence alignment of 16S rRNA gene sequences for the set of 29 strains (28 S. agalactiae and 1 E. coli) was performed with CLUSTALW algorithm integrated in MEGA [20]. Neighbor-Joining phylogenetic tree was reconstructed with Jukes-Cantor model [21]. The topology of phylogenetic tree was considered reliable upon 1000 replication of each sequence (Bootstrap value=1000) by elimination of nucleotide positions with gaps and missing data.
Statistical Analysis
Entry of collected data and statistical analyses were conducted using the SPSS software version 16.0. Chi-square test and Fischer’-exact test (wherever applicable) was used for statistical significance with p≤0.05; Confidence Interval (CI)-95%.
Results
A total number of 345 vaginal swabs were obtained from pregnant women at 35-37 weeks of gestation. Out of the total 345 pregnant women, 52 (15.1%) were found to be colonised with GBS and 293 (84.9%) were found to be non-colonised with GBS. Preliminary identification of GBS was done by gram staining [Table/Fig-1] and further confirmed through biochemical tests.
Amongst GBS positive cases, women belonging to less than 25 years of age (51.9%, n=27/52) was seen to be GBS colonised at a higher rate [Table/Fig-2]. Whereas literate women were found to be positive at a rate of 80.7% (n=42/52). On the other hand, more cases were observed in economically weaker section (92.3%, n=48/52). Most of the GBS colonised women in their gestation period had no Urinary Tract Infection (UTI) at a rate of 84.6% (44/52) and 15.4% (8/52) had presented with UTI. While 88.5% (n=46/52) of them had no past history of UTI and 11.5% (6/52) of them suffered earlier from UTI. Women with vaginal discharge was seen to be colonised with GBS at a rate of 30.8% (n=16/52) where 26.9% (14/52) had white colour discharge, 1.9% (1/52) had yellowish discolouration and 1.9% (1/52) had clear discharge. Mild odour of vaginal discharge was present in 1.9% (1/52) of the colonised women. Vaginal itching was present in 15.4% (8/52) of the colonised women. Colonisation rate was seen more in multigravida women (53.8%, n=28/52). Majority of the GBS colonised mothers delivered neonates vaginally (76.9%, n=40/52).
Sociodemographic and obstetric characteristics of GBS colonised women.
| Demographic and obstetrics details | Positive, N (%) | Negative, N (%) | Total | p-value |
|---|
| Age (years) |
| ≥25 | 25 (18.4%) | 111 (81.6%) | 136 | 0.166* |
| <25 | 27 (12.9%) | 182 (87.1%) | 209 |
| Qualification |
| Uneducated | 10 (17.9%) | 46 (82.1%) | 56 | 0.161* |
| Educated | 42 (14.5%) | 247 (85.5%) | 289 |
| Occupation |
| Unemployed | 10 (14.1%) | 61 (85.9%) | 71 | 0.480# |
| Employed | 42 (15.3%) | 232 (84.7%) | 274 |
| Income |
| ≤5000 | 48 (14.5%) | 284 (85.5%) | 332 | 0.116# |
| >5000 | 4 (30.8%) | 9 (69.2%) | 13 |
| Gravida |
| Primigravida | 24 (12.9%) | 162 (87.1%) | 186 | 0.143# |
| Multigravida | 28 (17.6%) | 131 (82.4%) | 159 |
| Vaginal discharge |
| Yes | 16 (20%) | 64 (80%) | 80 | 0.112# |
| No | 36 (13.6%) | 229 (86.4%) | 265 |
| Vaginal discharge colour |
| Clear | 1 (1.9%) | 6 (2.0%) | 7 | 0.312* |
| Yellowish | 1 (1.9%) | 1 (0.3%) | 2 |
| White | 14 (27%) | 57 (19.5%) | 71 |
| Absent | 36 (69.2%) | 229 (78.2%) | 265 |
| Vaginal discharge odour |
| Absent | 51 (14.9%) | 291 (85.1%) | 342 | 0.38# |
| Mild | 1 (33.3%) | 2 (66.7%) | 3 |
| Vaginal itching |
| Absent | 44 (13.5%) | 283 (86.5%) | 327 | 0.004* |
| Present | 8 (44.4%) | 10 (55.6%) | 18 |
| UTI in current pregnancy |
| Yes | 8 (21.1%) | 30 (78.9%) | 38 | 0.194# |
| No | 44 (14.3%) | 263 (85.7%) | 307 |
| Past UTI |
| Present | 6 (8.8%) | 62 (91.2%) | 68 | 0.13# |
| Absent | 46 (16.6%) | 231 (83.4%) | 277 |
| Type of delivery |
| Vaginal | 40 (16.7%) | 199 (83.3%) | 239 | 0.350* |
| CS | 12 (11.8%) | 90 (88.2%) | 102 |
| Lost contact | - | 4 | 4 |
| Maturity |
| Term | 49 (14.8%) | 282 (85.2%) | 331 | 0.238* |
| Preterm | 3 (37.5%) | 5 (62.5%) | 8 |
| LC/Home delivery | - | 6 | 6 |
| Gender of the child |
| Male | 28 (16.1%) | 146 (83.9%) | 174 | 0.822* |
| Female | 24 (14.4%) | 143 (85.6%) | 167 |
| LC | - | 4 | 4 |
*p-value obtained by Pearson chi-square test; #p-value obtained by Fisher’s-exact test; *GBS: Group B streptococcus; UTI: Urinary tract infection; LC: Lost contact; CS: Caesarean section
Most of the neonates of the colonised mothers were born at term stage (94.2%, n=49/52). Male neonates (53.8%, n=28/52) and female neonates (46.1%, n=24/52) were born to GBS positive patients. In this study, vaginal itching was found to be significantly associated (p=0.004; p≤0.05) with the increased rate of GBS colonisation while the other risk factors were not found to be significantly associated with mothers having GBS colonisation (Chi-square test-two-sided; Fisher’s-Exact test- wherever applicable).
Presence of scpB gene: The scpB gene was detected in all the 52 isolates with PCR amplified targeted product of size 255 base pairs.
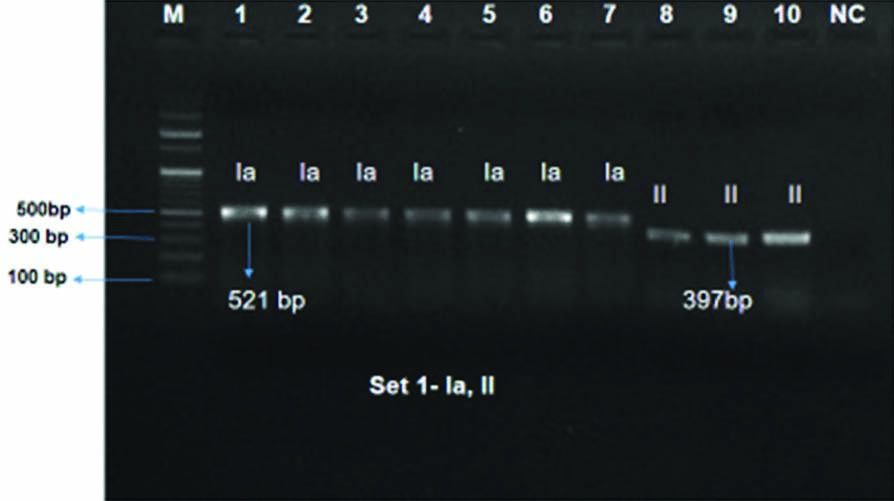
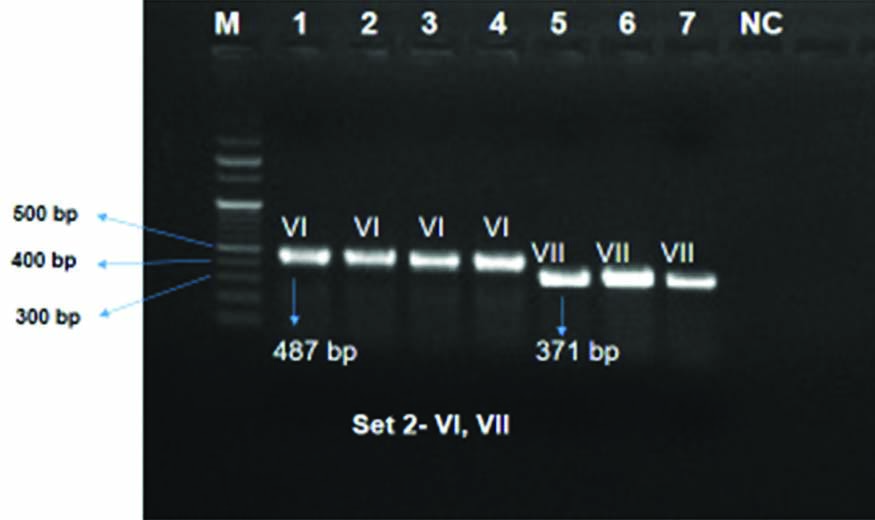
Serotyping of the GBS isolates: Out of the 52, a total of 30 GBS isolates could be revived and were taken for serotyping in the present study out of which 19 could be serotyped. The serotypes found in this study were Ia (42.1%, n=8/19), II (15.8%, n=3/19), VI (31.6%, n=6/19), VII (10.5%, n=2/19) [Table/Fig-3,4 and 5].
Serotype distribution of the GBS isolates. (Set 1)- Ia, Ib, II, III, IV; (Set 2)-V, VI, VII, VIII, IX
| Set 1- GBS serotypes (n, %) | Set 2- GBS serotypes (n, %) |
|---|
| Recruited pregnant women, GBS colonised (n=19) | Ia | Ib | II | III | IV | Recruited pregnant women, GBS colonised (n=19) | V | VI | VII | VIII | IX |
| 8 (42.1%) | Nil | 3 (15.8%) | Nil | Nil | Nil | 6 (31.6%) | 2 (10.5%) | Nil | Nil |
Antibiotic susceptibility pattern of the vaginal GBS isolates from pregnant women.
| Serotypes | Ia | II | VI | VII | NT |
|---|
| Antibiotics | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant |
| Cefotaxime | 7 (23.3%) | 1 (3.3%) | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 10 (33.3%) | 1 (3.3%) |
| Erythromycin | 6 (20%) | 2 (6.7%) | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 8 (26.7) | 3 (10%) |
| Clindamycin | 7 (23.3%) | 1 (3.3%) | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | - | 11 (36.7%) |
| Penicillin | 8 (26.7%) | - | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 11 (36.7%) | - |
| Ampicillin | 8 (26.7%) | - | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 11 (36.7%) | - |
| Ciprofloxacin | 8 (26.7%) | - | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 10 (33.3%) | 1 (3.3%) |
| Vancomycin | 8 (26.7%) | - | 2 (6.7%) | 1 (3.3%) | 6 (20%) | - | 2 (6.7%) | - | 11 (36.7%) | - |
*NT: Not typeable
Presentation of Capsular Polysaccharides (CPS) of GBS positive isolates by multiplex PCR Set 1: lane: 1-7: serotype Ia, lane: 8-10: serotype II, Lane M: 100 bp ladder, NC: Negative control.
Presentation of Capsular Polysaccharides (CPS) of GBS positive isolates by multiplex PCR Set 2: lane: 1-4: serotype VI, lane: 5-7: serotype VII, Lane M: 100 bp ladder, NC: Negative control.
Antibiotic susceptibility of the GBS serotypes: Analysis of antibiotic susceptibility test of the retrieved serotypes depicted that resistant to cefotaxime (3.3%, 3.3%), erythromycin (6.7%, 10%) and clindamycin (3.3%, 13.33%) was strictly shown by serotype Ia and Not Typeable (NT) GBS respectively. While 3.3% isolates from both serotype II and NT showed resistant to vancomycin and ciprofloxacin respectively [Table/Fig-6].
Antibiotic susceptibility pattern of the vaginal GBS isolates from pregnant women.
| Serotypes | Ia | II | VI | VII | NT |
|---|
| Antibiotics | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant |
| Cefotaxime | 7 (23.3%) | 1 (3.3%) | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 10 (33.3%) | 1 (3.3%) |
| Erythromycin | 6 (20%) | 2 (6.7%) | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 8 (26.7) | 3 (10%) |
| Clindamycin | 7 (23.3%) | 1 (3.3%) | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | - | 11 (36.7%) |
| Penicillin | 8 (26.7%) | - | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 11 (36.7%) | - |
| Ampicillin | 8 (26.7%) | - | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 11 (36.7%) | - |
| Ciprofloxacin | 8 (26.7%) | - | 3 (10%) | - | 6 (20%) | - | 2 (6.7%) | - | 10 (33.3%) | 1 (3.3%) |
| Vancomycin | 8 (26.7%) | - | 2 (6.7%) | 1 (3.3%) | 6 (20%) | - | 2 (6.7%) | - | 11 (36.7%) | - |
*NT: Not typeable
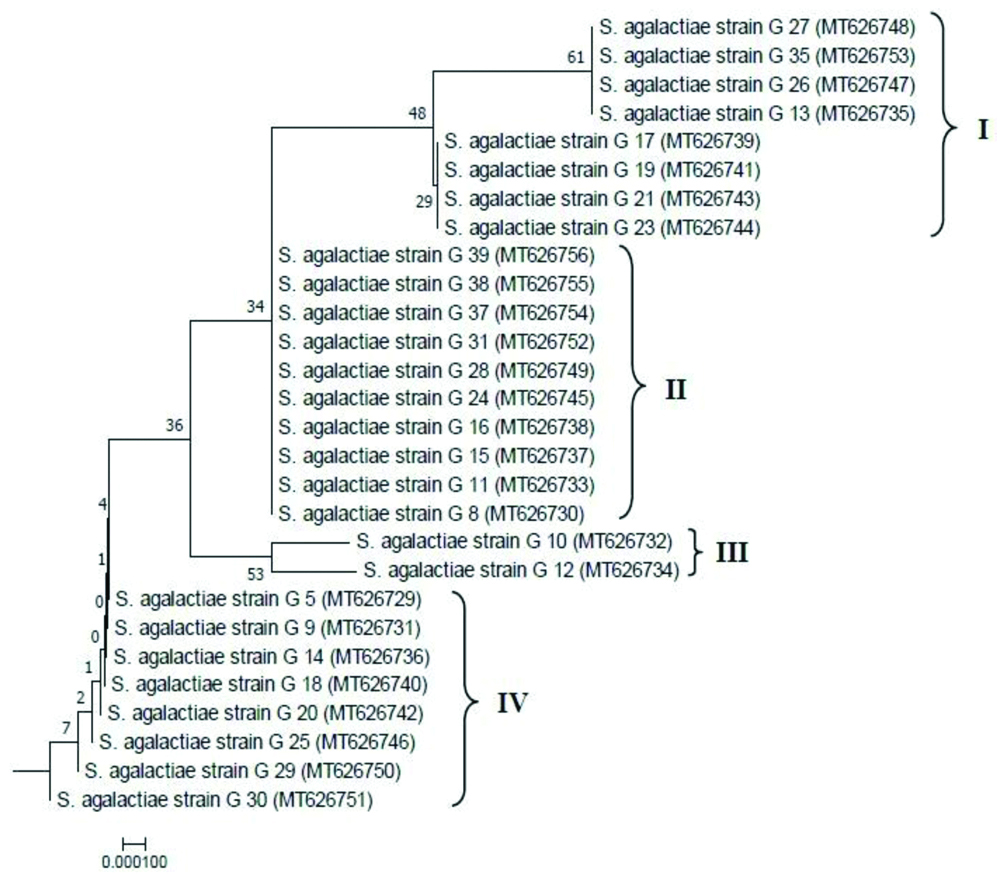
Phylogenetic analysis of the GBS isolates: In the reconstructed neighbor-joining tree, E.coli was shown as an out group from the group of S.agalactiae strains [Table/Fig-7a]. The optimal condensed tree is presented with the sum branch length=0.34425054. The sub-tree of S.agalactiae group (separated from the main tree) is shown expanded in [Table/Fig-7b]. Based on the nucleotide diversity in the 16S rRNA partial gene sequences, S.agalactiae strains in this study were found to be grouped into four distinct clusters designated as I, II, III and IV in the sub-tree with distinct phylogenetic relationship. As observed the variations among the isolates in [Table/Fig-7b], Cluster II comprising 10 isolates (out of 28) in the phylogenetic tree was observed to be the major cluster depicting that the frequent S. agalactiae isolates circulating in this part of the study site may be grouped majorly under cluster II. Additionally, isolates under cluster I and IV (having eight isolates in each cluster) were also found to be less frequent in comparison to the isolates under cluster II.
Reconstructed Neighbor-Joining phylogenetic tree based on 16S rRNA gene sequences with evolutionary distances computed by Jukes-Cantor model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Taxa in the tree presented by their strain names and GenBank accession numbers in parentheses.
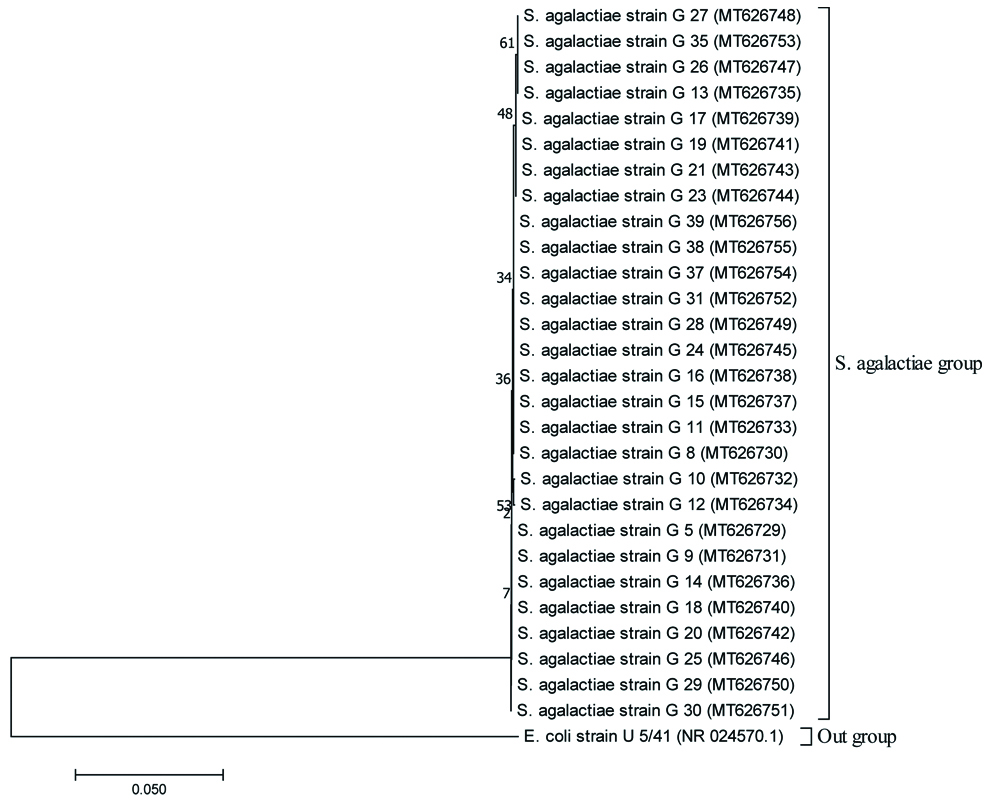
Sub-tree of S. agalactiae group expanded from the main tree (Fig. 4a) representing 4 clusters viz., cluster I, II, III, IV.
Discussion
Vaginal niche colonised by GBS in expectant mothers have developed a worrisome concern for the clinicians in terms of determining neonatal healthy outcome. This matter in infants has pressurised to investigate as newborns midst their birth gets exposed to maternal GBS. Very few South-Asian countries have reported on GBS with varying prevailing rates recording a comprehensive colonisation rate of 13% [22]. Screening pregnant women in the last trimester is mandatory as recommended by CDC to thwart infants being victim to encroaching GBS [23]. This hospital based study from Dibrugarh, Assam of northeast India, consequently has grabbed the attention to reveal GBS status in late trimester women of the region. In the present study, a carriage rate of 15.1% has been found out akin to the earlier Indian study done by Chaudhary M et al., [5]. Another report from Srilanka has shown similar findings at the boundary rate of 14.8% [24]. GBS prevalence found in this study was quite high in comparison to the prior studies done in different places in India [3,25-33]. These few studies from India on pregnant women have thrown some light on GBS prevailing rate although difference in the sample size, site of collection, culture techniques, molecular methods for which a variation in the rates can be seen. However, the rate of colonisation in present study was lower than Pakistan (17%) [34].
The fluctuation of the prevailing rates of GBS infection in pregnant women necessitates further research to capture a precise rate following a mandatory universal standard guideline [5,6]. These studies have demonstrated the rate of Streptococcus agalactiae amidst pregnant mothers ranging from 1.62-15% across different regions [5,6]. Other parts of the countries reported a higher rate of colonisation in North America (18.3%) [35], South Africa (37%) [14], Zimbabwe (31.6%) [36].
CPS of Streptococcus agalactiae possesses its virulence property which causes invasive infection in neonates from its colonised mother. Therefore, targeting CPS vaccines can be an effective measure in treating GBS colonised patients. None of the Indian studies have reported molecular based serotypes but have declared based on latex agglutination test. In the current study, serotyping has been performed in 30 isolates from GBS positive mothers. To the best of our knowledge, this is the first study on Streptococcus agalactiae in pregnant women from Assam, India. Study report of such kind has not been reported from this part of the country detailing a comprehensive picture of GBS serotypes in colonising pregnant women, their susceptibility to antibiotics and existing phylogenetic relatedness. However, serotypes viz., III, V, and Ib which have been found by latex agglutination test in Delhi are not observed among pregnant women through molecular method followed in the present study [5].
In the current study, the dominant serotype was Ia followed by VI, II and VII. Compared to serotype III which is more prevalent in China [37], other Asian countries have reported the higher rate of distribution of serotypes VI, VII, VIII and IX [22]. Serotype III is completely absent in present study. The dominant serotype Ia followed by VI in this study site are similar amongst the 5 global lofty serotypes [22]. In a different site within this country, serotype III has been seen to be dominant accompanied by serotypes V, II, Ia, VII and Ib based on latex test method [5]. Although differential observations for GBS serotypes have been recorded from the reporting studies, additional research in this context is needed to shed light on the prevailing serotypes throughout the country [5,22]. It is worth mentioning that a previous study from Assam has found GBS in cerebrospinal fluid from one neonate born vaginally to GBS colonised mother [8].
Maternal administration of penicillin with alternative to ampicillin is recommended by ACOG (American College of Obstetricians and Gynaecologists) for preventing neonatal GBS as it is categorised in the first line therapy by the clinicians [4]. For lower penicillin allergic response women, cephalosporin (particularly cefotaxime) group of antibiotics is referred while clindamycin to higher allergic group of GBS positive mothers. Intrapartum antibiotic therapy for healthy outcome of the newborns thus is mandatory for the GBS colonised women. As evident from this study about the complete susceptibility of the GBS isolates to penicillin and ampicillin collectively from both serotyped (Ia, II, VI, VII) and not typeable GBS, resistance to cefotaxime, erythromycin, clindamycin by serotype Ia, vancomycin by serotype II has also been documented. Moreover, resistance is also persistent among the not typeable GBS against cefotaxime, erythromycin, clindamycin and ciprofloxacin.
As the antimicrobial resistant profile by serotypes is observed, it is seen that serotype Ia and not typeable isolates showed resistant to cefotaxime, erythromycin and clindamycin. Similar report by other Indian studies on complete sensitivity of GBS towards penicillin and ampicillin has been seen [25,26,28,31,33,38]. The findings of higher rate of resistance in GBS to erythromycin and clindamycin in present study is in consistent with earlier reports [28,31,38]. With such an increasing rate of resistance in GBS to penicillin allergic alternatives (clindamycin and erythromycin), a blueprint comprising the prevalent serotypes is the utmost solution to tackle this global plight. Previous study on human derived GBS strains across different countries explained a total of five major clone complexes differ from bovine clone complexes [39]. Pertaining to phylogeny, these major clone complexes along with a new clone complex (viz., CC103 that infects both human and bovine) have distinct phylogenetic relationship [40]. Consistent to this, the neighbour joining phylogeny result obtained in this study have shown four distinct clusters, each of them has unique evolutionary branches indicative of existing nucleotide diversity in the human derived GBS population of the selected study area.
Limitation(s)
This study, for the first time, brought the picture of GBS infection in antepartum women. It has comprehensively focused the prevailing GBS serotypes. However, the study would have been more profound if the analysis would include swab samples from both lower vaginal and rectal region of the subjects, which would have brought us a complete picture of actual burden rate of Streptococcus agalactiae and its circulating serotypes in expectant mothers.
Conclusion(s)
In the field of health sector, status of GBS burden amongst pregnant mothers is still unknown particularly in Assam, India. But the very low rate of neonatal morbidity has put up the fact that extensive care is carried out in the neonatal units of the healthcare settings. The present study has highlighted the prevailing GBS scenario which may be helpful in declining the rate of adverse neonatal outcomes with on time maternal therapeutic administration and routine antibiogram profiling. Moreover, detail profile of serotypes based on CPS of GBS may pave a way of designing vaccines for immunising the expectant mothers to prevent adverse outcomes of the newborns which would contribute in out rooting the burden, either partially or completely, from the population in coming time.
*p-value obtained by Pearson chi-square test; #p-value obtained by Fisher’s-exact test; *GBS: Group B streptococcus; UTI: Urinary tract infection; LC: Lost contact; CS: Caesarean section
*NT: Not typeable
*NT: Not typeable