Pulmonary tuberculosis is life threatening disease and worldwide it is second only to Human Immunodeficiency Virus (HIV) infection. TB is one of the top 10 causes of death and the leading cause from a single infectious agent (above HIV/AIDS). In 2019, an estimated 10 million people fell ill with TB worldwide including 1.2 million children and among them 1.4 million people died. TB is present in all countries and at every age groups [1]. Adding to the considerable burden of TB associated complications are rapid emergence of in drug resistant strains of mycobacterium tuberculosis and developing countries account for more than 90% cases worldwide [2].
Early diagnosis and treatment is essential for the control of the spread of pulmonary TB. However, it is practically very difficult disease to do laboratory confirmation. The gold standard of diagnosis of Mycobacterium tuberculosis is growth on Lowenstein Jensen media but culture requires 4-6 weeks long incubation time due to slow growth of Mycobacteria [3]. Sputum smears staining with Ziehl-Neelsen staining followed by bright field microscopy is widely used in clinical laboratories but it has low sensitivity and specificity. Polymerase Chain Reaction (PCR) based amplification assays have revolutionised the TB diagnostics however, inhibition of reaction by nonspecific factors leads to cautious interpretation in clinical use. Serological test can be used but are time consuming and cross reacting antibodies lead to interpretation problem [4,5]. New biomarker for diagnosis of pulmonary tuberculosis are needed.
Recently, miRNAs has been intensively studied as diagnostic and prognostic markers for chronic non-infectious diseases like cardiovascular disease, malignancy, diabetes mellitus, neurological illness etc. MicroRNAs are small noncoding RNAs that plays a key role in maintenance of various biological process, such as immunoregulation, organogenesis, tumorigenesis. They are small conserved nucleotides and are 18-22 nucleotide long. These tiny miRNAs are vital components of new approach of post-transcriptional regulation of gene expression. A total of more than 2000 miRNAs have been reported in human genome [6-8].
Extracellular miRNA are stable and as they are not affected by intracellular endogenous RNAses. Recently, studies have come exploring the diagnostic potential of circulating miRNA for malignancies and infectious diseases in biological fluids like whole blood, plasma, serum etc., [9-11]. This study was planned with primary objective to identify serum miRNAs differentially expressed in pulmonary tuberculosis patients and evaluate the use of miRNA as biomarker for diagnosis of pulmonary tuberculosis.
Materials and Methods
This prospective cross-sectional laboratory based study was conducted from 1st August, 2012 to 7th July, 2018. The patients were recruited at Lala Lajpat Rai Hospital, Kanpur, Uttar Pradesh, India. Conventional test were performed at Department of Microbiology, GSVM Medical College, Kanpur, Uttar Pradesh, India, and miRNA related molecular work was performed at Department of Microbiology, Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow. The study protocol was approved by the Institute’s Ethical Committee by (Letter Number 86/IEC/Principal Steno/11) and written Informed consent was collected from all cases and controls before recruiting them.
Sample size calculation: To compare the dysregulated miRNA among pulmonary TB cases and controls, considering 2 miRNA that will be dysregulated, at minimum two sided 95% confidence interval and 80% power of the study, estimated sample size for cases and control was 35 each (total 70 patients) [12]. Sample size was estimated using software power analysis and sample size version-13.0 (PASS-13, NCSS, LLC, USA).
Cases and Control Recruitment
Acute pulmonary TB cases were examined clinically and underwent PCR for insertion sequence 6110 of Mycobacterium tuberculosis on sputum sample. PCR results positive cases were included in study and healthy controls not suffering from pulmonary tuberculosis along with negative Interferon gamma release assay were recruited as control. Only active cases of pulmonary TB were included in study. Chronic or healed cases of pulmonary TB were excluded from study.
TaqMan MicroRNA Array Quantitative PCR
A 2 mL of venous blood was collected from each participant using aseptic precautions. RNA was isolated from blood using miRNA mini Kit (Qiagen, Hilden, Germany) as per kit instructions. The TaqMan MicroRNA Reverse Transcription Kit (Thermofischer, USA) was used for preparation of complimentary DNA. A reverse transcriptase reaction was done on a CFX 96 (Biorad, USA) with the following conditions: 45°C for 20 minutes; 94°C for 5 minutes and holding at 4 degree. Human TaqMan low density miRNA array (Applied Biosystems, USA) was performed on five acute pulmonary TB cases and five healthy controls. The assay enables accurate quantitation of 754 human micro RNAs simultaneously and identifies the miRNAs. Differential expressed in pulmonary tuberculosis patients. To identify these miRNA following criteria was used; Cycle Threshold (CT) values should be less than 35 and miRNA levels difference between the patient and control groups should be ≥2 fold [10].
Quantification of Identified Candidate miRNA by Real Time PCR
TaqMan miRNAs Array results were further validated via quantification of targeted microRNAs in 30 cases and 30 controls by TaqMan Real time PCR (Applied Biosystems, USA). Reverse transcriptase reactions were performed using the ABI miRNA reverse transcription kit as per manufacturer’s instructions. The PCRs were carried out with seven minutes incubation at 94°C followed by 45 cycles of 94°C for 10 seconds and 58°C for one minute in a reaction volume of 25 μL using an ABI 7600 RT-PCR System. A typical reaction consisted of 5 μL diluted cDNA, 12.5 μL TaqMan Universal PCR Master Mix (2X) and 0.5 μL TaqMan miRNA Assay primer-probe (Applied Biosystems) and 7 μL nuclease free water. Each sample was run in triplicate. Cut off threshold value was taken as cycle number of RTPCR at which the fluorescence emitted by FAM probe passed the baseline threshold. The results were normalised with miRNA-16 as an endogenous control [11-13].
Statistical Analysis
Data was collected and evaluated with SPSS software version 16.0 (SPSS, Inc., Chicago, USA) and presented as means±standard deviations. Student’s t-tests and Analysis of Variance (ANOVA) tests were used for statistical analysis. The p-value of less than 0.05 was considered statistically significant. The ROC curve were generated using MedCalc data analysis software and Area Under Curve (AUC) were calculated to determine the statistical significance of dysregulated miRNA in cases of pulmonary tuberculosis [14].
Results
Demographic Profile of Cases and Controls
A total of 96 suspected cases of acute pulmonary TB were examined clinically and underwent PCR testing on sputum sample. Results showed that 35 cases were positive for Mycobacterium tuberculosis and were recruited as cases. Of these recruited patients, 18 were males and 17 were females and their mean age was 32 years (21-40 years). Thirty five age and sex-matched healthy controls were also recruited.
MiRNA Profiling using TaqMan Low Density Array miRNA Analysis (TLDA)
The test was performed on 5 cases and 5 controls; results showed that 48 miRNAs were upregulated and 27 were downregulated in pulmonary TB cases. miRNA expression was increased in cases when compared to control on average by 3.05 to 210 fold but decreased up to 0.39-fold [Table/Fig-1,2]. Upregulated and downregulated miRNA are shown in heat map chart [Table/Fig-3,4]. Among these, 5miRNAs (hsa-miRNA-29a, hsa-miRNA 19a, hsa-miRNA-519c, hsa-miRNA-384, hsa-miRNA 105) were significantly upregulated and 2 miRNAs (hsa- miRNA-377 and hsa-miRNA-504) were significantly downregulated (≥5 CT difference between the patient and control groups). These miRNAs were selected for further evaluation via RT-PCR on 30 TB infected cases and 30 healthy controls.
Upregulated miRNAs in tuberculosis patients.
| Name | Active TB/Controla |
|---|
| hsa-miR-576-5p | 3.05 |
| hsa-miR-101 | 3.05 |
| hsa-miR-302b | 3.18 |
| hsa-miR-4308 | 3.32 |
| hsa-miR-574-3p | 3.37 |
| hsa-miR-208b | 3.62 |
| hsa-miR-1184 | 3.81 |
| hsa-miR-500 | 3.87 |
| hsa-miR-125a-5p | 3.95 |
| hsa-miR-744 | 4.11 |
| hsa-miR-296 | 4.11 |
| hsa-miR-3911 | 4.31 |
| hsa-miR-4279 | 4.51 |
| hsa-let-7i | 4.71 |
| hsa-miR-487b | 4.74 |
| hsa-miR-19a | 4.8 |
| hsa-miR-628-5p | 4.82 |
| hsa-miR-103-as | 4.83 |
| hsa-miR-4285 | 5.03 |
| hsa-let-7c | 5.22 |
| hsa-miR-381 | 5.22 |
| hsa-miR-199a | 5.37 |
| hsa-miR-1915 | 5.38 |
| hsa-miR-218-2-3p | 5.42 |
| hsa-miR-379 | 5.45 |
| hsa-miR-1284 | 5.45 |
| hsa-miR-4287 | 5.56 |
| hsa-miR-486-3p | 6.31 |
| hsa-miR-183 | 6.38 |
| hsa-miR-22 | 7.41 |
| hsa-miR-146b | 7.42 |
| hsa-miR-502-3p | 7.44 |
| hsa-miR-365 | 7.52 |
| hsa-miR-375 | 8.15 |
| hsa-miR-93 | 8.24 |
| hsa-miR-382 | 8.61 |
| hsa-miR-9 | 8.67 |
| hsa-miR-24 | 8.72 |
| hsa-miR-125b | 11.2 |
| hsa-miR-31 | 11.4 |
| hsa-miR-519c | 11.6 |
| hsa-miR-103 | 24.7 |
| hsa-miR-1280 | 11.6 |
| hsa-miR-29a | 12.7 |
| hsa-miR-3606 | 20 |
| hsa-miR-105 | 21 |
| hsa-miR-1470 | 22.3 |
| hsa-miR-548b-5p | 23.2 |
| hsa-miR-384 | 210 |
aExpression ratio of TB serum/control serum
Downregulated miRNAs in tuberculosis patients.
| Name | Active TB/Controla |
|---|
| hsa-miR-377 | 0.02 |
| hsa-miR-504 | 0.12 |
| hsa-miR-221 | 0.13 |
| hsa-miR-944 | 0.18 |
| hsa-miR-3153 | 0.19 |
| hsa-miR-205 | 0.19 |
| hsa-miR-3145 | 0.20 |
| hsa-miR-4311 | 0.22 |
| hsa-miR-181 | 0.22 |
| hsa-miR-758 | 0.23 |
| hsa-miR-154 | 0.23 |
| hsa-miR-590-5p | 0.24 |
| hsa-miR-519d | 0.25 |
| hsa-miR-769-5p | 0.26 |
| hsa-miR-222 | 0.27 |
| hsa-miR-380 | 0.30 |
| hsa-miR-512-3p | 0.30 |
| hsa-miR-618 | 0.30 |
| hsa-miR-186 | 0.32 |
| hsa-miR-337-5p | 0.32 |
| hsa-miR-3666 | 0.32 |
| hsa-miR-3125 | 0.32 |
| hsa-miR-518d-5p | 0.33 |
| hsa-miR-3612 | 0.33 |
| hsa-miR-374 | 0.34 |
| hsa-miR-371-3p | 0.36 |
| hsa-miR-320 | 0.39 |
aExpression ratio of TB serum/control serum
Heat sensitive mapping showing upregulated miRNAs in tuberculosis patients.
| Name | Acute tuberculosis cases/Healthy control |
|---|
| hsa-miR-384 | 210 |
| hsa-miR-103 | 24.7 |
| hsa-miR-548b-5p | 23.2 |
| hsa-miR-1470 | 22.3 |
| hsa-miR-105 | 21 |
| hsa-miR-3606 | 20 |
| hsa-miR-29a | 12.7 |
| hsa-miR-519c | 11.6 |
| hsa-miR-31 | 11.4 |
| hsa-miR-125b | 11.2 |
| hsa-miR-24 | 8.72 |
| hsa-miR-9 | 8.67 |
| hsa-miR-382 | 8.61 |
| hsa-miR-93 | 8.24 |
| hsa-miR-375 | 8.15 |
| hsa-miR-365 | 7.52 |
| hsa-miR-502-3p | 7.44 |
| hsa-miR-146b | 7.42 |
| hsa-miR-22 | 7.41 |
| hsa-miR-183 | 6.38 |
| hsa-miR-486-3p | 6.31 |
| hsa-miR-4287 | 5.56 |
| hsa-miR-379 | 5.45 |
| hsa-miR-1284 | 5.45 |
| hsa-miR-218-2-3p | 5.42 |
| hsa-miR-1915 | 5.38 |
| hsa-miR-199a | 5.37 |
| hsa-miR-381 | 5.22 |
| hsa-let-7c | 5.22 |
| hsa-miR-4285 | 5.03 |
| hsa-miR-103-as | 4.83 |
| hsa-miR-628-5p | 4.82 |
| hsa-miR-19a | 4.8 |
| hsa-miR-487b | 4.74 |
| hsa-let-7i | 4.71 |
| hsa-miR-4279 | 4.51 |
| hsa-miR-3911 | 4.31 |
| hsa-miR-744 | 4.11 |
| hsa-miR-296 | 4.11 |
| hsa-miR-125a-5p | 3.95 |
| hsa-miR-500 | 3.87 |
| hsa-miR-1184 | 3.81 |
| hsa-miR-208b | 3.62 |
| hsa-miR-574-3p | 3.37 |
| hsa-miR-4308 | 3.32 |
| hsa-miR-302b | 3.18 |
| hsa-miR-576-5p | 3.05 |
| hsa-miR-101 | 3.05 |
Heat sensitive mapping depicting downregulated miRNAs in tuberculosis patients.
| Name | Acute tuberculosis cases/Healthy control |
|---|
| hsa-miR-320 | 0.39 |
| hsa-miR-371-3p | 0.36 |
| hsa-miR-374 | 0.34 |
| hsa-miR-518d-5p | 0.33 |
| hsa-miR-3612 | 0.33 |
| hsa-miR-186 | 0.32 |
| hsa-miR-337-5p | 0.32 |
| hsa-miR-3666 | 0.32 |
| hsa-miR-3125 | 0.32 |
| hsa-miR-380 | 0.3 |
| hsa-miR-512-3p | 0.3 |
| hsa-miR-618 | 0.3 |
| hsa-miR-222 | 0.27 |
| hsa-miR-769-5p | 0.26 |
| hsa-miR-519d | 0.25 |
| hsa-miR-590-5p | 0.24 |
| hsa-miR-758 | 0.23 |
| hsa-miR-154 | 0.23 |
| hsa-miR-4311 | 0.22 |
| hsa-miR-181 | 0.22 |
| hsa-miR-3145 | 0.2 |
| hsa-miR-3153 | 0.19 |
| hsa-miR-205 | 0.19 |
| hsa-miR-944 | 0.18 |
| hsa-miR-221 | 0.13 |
| hsa-miR-504 | 0.12 |
| hsa-miR-377 | 0.02 |
Quantitative RT-PCR Analysis of Dysregulated miRNA
Quantitative RT-PCR performed on 30 cases and matched control each targeting seven candidate miRNAs (5 upregulated and 2 downregulated) showed that expression level of miRNA 29a in pulmonary TB cases was significantly upregulated followed by miRNA 384 when compared with expression level of healthy controls. Further, miRNA 504 was significantly downregulated in TB cases as compared to controls. There was no significant differences detected in the expression of hsa-miRNA- 519c, hsa-miRNA- 19a, hsa-miRNA-105 and hsa-miRNA- 377 (p>0.05).
Receiver Operating Characteristic (ROC) Analysis
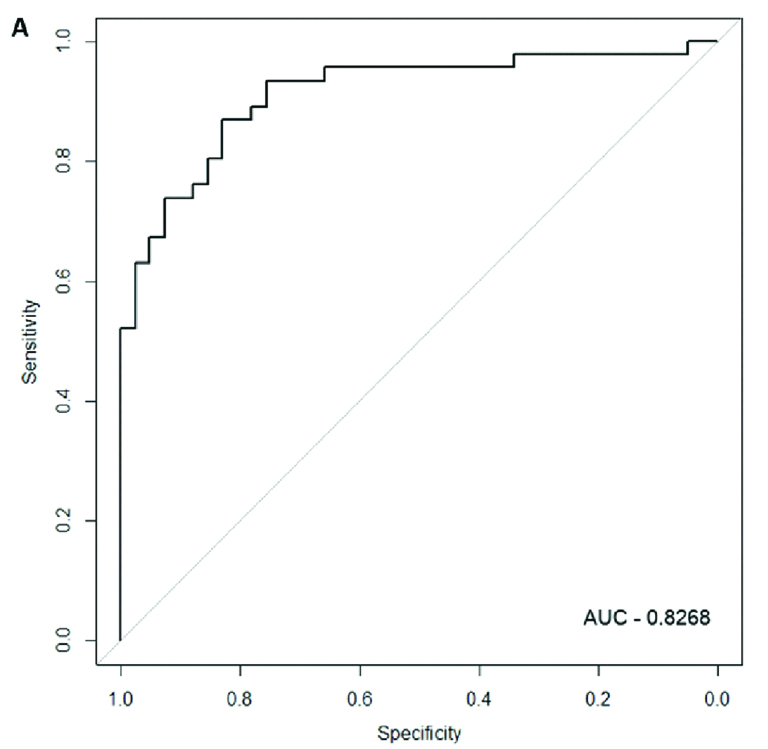
Expression data for over expressed miRNA in cases and control groups were used to build an ROC plot and found that AUC of miRNA-29a was 0.8268 (sensitivity=80%, specificity=82%) which reflected reasonable separation between the two groups. Thus circulating miRNA-29a could be used as a diagnostic biomarker for active pulmonary TB [Table/Fig-5].
ROC curve of serum miR-29a to differentiate pulmonary TB cases from controls (Sensitivity: 80% and Specificity: 82%).
Discussion
MicroRNAs are conserved nucleotides working at post transcriptional level and are responsible for regulation of biological functions of eukaryotic cell. The capability of organisms to rapidly adapt to the changing environment is essential for its survival and miRNAs plays an important role in cells to rapidly transfer and internalise an external signal. In addition to its action on translation, the miRNAs also affects epigenetic processes. miRNAs appear to target about 60% of the genes of humans and other mammals and have been identified in almost all kingdoms of life, including Achaea, humans and plants [15]. However, the way in which miRNA regulates the expression of mRNA at the translational level in TB remains to be elucidated.
Recently several new studies have explored the role of miRNA as diagnostic biomarker in cases with active pulmonary tuberculosis. Multiple microarrays have been conducted on wide range of clinical specimens like blood mononuclear cells, pleural fluid, plasma, serum, bronchoalveolar lavage, sputum, and even mononuclear cells stimulated with mycobacterial antigens ex-vivo and found differential expression of specific miRNAs in their studies [16-20].
Liu Y et al., studied miRNA dysregulated in cases of pulmonary tuberculosis and reported that miRNA-144 was significantly upregulated in blood of pulmonary TB patients [16]. Fu Y et al., studied the free extra cellular circulating miRNAs in pulmonary tuberculosis patients and reported that 59 miRNAs were upregulated and 33 downregulated; miRNA-93 was maximum up regulated and miRNA-518d-5p, miRNA-520c-5p and miRNA-526a were the most downregulated miRNAs [13]. Guo W et al., performed a in silico software miRanda based study and predicted 26 M. tuberculosis genes associated with human miRNAs dysregulation [21].
In this study, it was observed that serum miRNA-29a levels were significantly higher in acute pulmonary TB patients compared with healthy controls (p=0.03). Further ROC analysis showed that miRNA-29a demonstrate a good distinguishing efficiency in discriminating the TB cases from the healthy subjects (AUC=0.8268; sensitivity=80%; specificity=82%). There was no statistical significant difference in expression of remaining miRNAs analysed between cases and controls. Several other workers have also reported that miRNA-29a are over expressed in cases of pulmonary TB [13,22]. Fu Y et al., reported similar findings and showed that area under ROC of miRNA-29a as 0.83, which shows that it has a great potential to act as biomarker for detection of pulmonary tuberculosis with good sensitivity and specificity of 83% and 80% respectively [13]. Role of miRNA in pathogenesis of TB and its interaction with IFN, gamma has also been elucidated in many studies. Similar to our study results another recent study also documented that miRNA29a are upregulated after the mycobacterium tuberculosis infection of human macrophages and this overexpression is associated with protective cytokine regulation [23,24]. Another study showed that miRNA-29a expression depended on sex, where Peripheral Blood Mononuclear Cell (PBMC) from females with active TB exhibited a lower expression than PBMCs of male patients however in this study, miRNA 29a expression was equal in both sexes [25].
All these studies regarding pathogenesis, interaction with IFN gamma and apoptosis suggest that miRNA-29a overexpression act as down regulator of immune response against mycobacterium tuberculosis infection and shows the potential of further research work on micro RNA modulation for prevention and therapy against mycobacterial infection. Till now there are many studies done in developed countries to evaluate role of miRNA-29a in pulmonary infection [12,18]. This study was first of its kind to emphasise expression of miRNA-29a in population of developing country like India.
Limitation(s)
The number of cases included in this study is small, so multi-centric study including larger numbers of pulmonary tuberculosis patients and healthy controls are required to validate our findings. Furthermore, only dysregulated miRNAs were evaluated and their biopathway were not explored.
Conclusion(s)
Current diagnostic techniques for diagnosing pulmonary TB have many shortcomings. This study results suggests that serum MiRNA-29a level can be used as an effective biomarker for diagnosis of pulmonary tuberculosis patients.