Introduction
Cytogenetically, human ‘Y’ chromosome is an acrocentric type composed of two Pseudoautosomal Regions (PAR1 and PAR2), a short arm (Yp) and long arm (Yq), that are separated by a centromere [1]. PARs and short arm Yp are euchromatic while a large portion of the long qarm is heterochromatic except around centromere which is euchromatic in nature. The non-recombining region (NRY) is the locus beyond the PARs which does not recombine with X-Chromosome during meotic paring [2]. This region has heterochromatic and euchromatic loci. The heterochromatic locus of Yq comprises distal Yq containing two highly repetitive sequences DYZ1 and DYZ2 [1,3]. Variation in this region of the long arm of Y has been observed [2], whose significance is much unknown [Table/Fig-1] [4].
Old and new classification of Y chromosome in relation to AZF with modification [4].
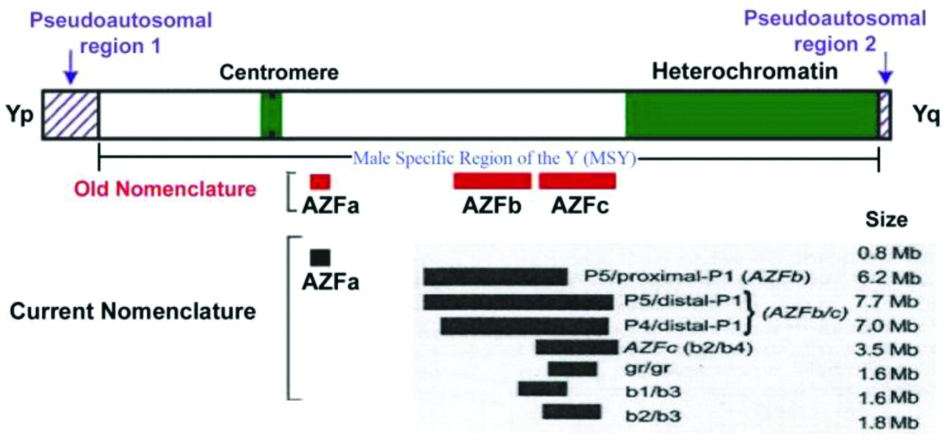
The euchromatic region encompasses the pericentromeric region and the short and long arms of Y which refer to the male specific region on Y (MSY; [Table/Fig-1]). This region contains genes for sex determination and regulation of brain functions [1,4,5]. These genes are critical in development of gonadal differentiation, development and function like Sex-Determining Region on Y Chromosome (SRY), Testis-Specific Protein on Y chromosome (TSPY), while the longarmed (Yq) has 12 genes and gene families mainly [1,5,6]. After this discovery, the Y chromosome is well studied in various species and several functional genes (approximately 70) are detected [7]. Basically these genes fall in two categories viz., housekeeping genes homologous to X and gene families expressed specific to testes, the candidate genes. The involvement of non-recombining region of Y(NRY) locus related to Yq in male infertility was coined in 1997, almost 23-year-ago. However, in 1976, Italian researchers identified detectable deletions at distal end of band Yq11 in 6 infertile males (6/1170). Their investigation revealed that deletions are de novo and could be related to azoospermia [8]. Based on this, the authors proposed aspermatogenesis factor called the AZF on the Yq locus. With the advent of molecular maps of the ‘Y’, the studies in AZF region and male infertility were extensively elaborated [9-11]. Using a panel of markers, infertile males who had deletions in the Yq 11.23 were identified [1,12,13]. These studies disclosed in this region, as three sub regions; proximal, middle and distal defined as AZFa, AZFb and AZFc, respectively [14]. The Deleted in Azoospermia (DAZ) gene in AZFc locus was considered as a strong candidate for male infertility, little is known about other genes in this locus [12]. This region has the size 0.8 Mb (AZFa), 6.2 Mb (AZFb/P5/Proximal-P1), 7.7 Mb (AZFb/c; P5/distal P1 and P4/distal P1), 3.5 Mb (AZFc (b2/b4), 1.6 Mb (gr/gr), 1.6 Mb (b1/b3) and 1.8 Mb (b2/b3) as mentioned in [Table/Fig-1] [1,15,16].
AZFa encodes only single copy genes of four mapped. These genes are X-homologous genes that escape during inactivation. These are ubiquitin peptidase 9 Y-linked (USPY9) whose function is beyond regulation of sperm production. Dead Box on Y Chromosome (DBY) deletion leads to Sertoli Cell Only Syndrome (SCOS) and involves also early stages of development Ubiquitously Transcribed Tetratricopeptide Repeat on Y Chromosome (UTY) gene status in male infertility is unknown. Same is for thymosine beta 4 Y linked (Tß4Y). Thus, these candidate genes contribute 5% of deletion [16-18]. AZFb genes contribute about 16% q deletions in infertile males. Deletions bring about azoospermia, oligospermia. It also contains admixture of AZFc locus [Table/Fig-1]. It encodes 5 single copy of genes and others are more than single copy. These are Cyorf15, RPS4Y2, E1F1AY, SMCY, XKRY, HSFY, PRY and RBMY. The RBMY is the most important genes of AZFb involved in spermatogenesis. Deletion of PRY and RBMY lead to hypospermatogenesis/complete loss of spermatogenesis [19-23]. Especially, RBMY protein is involved in all stages of sperm production and male cancers [12]. AZFc locus contributes to 60% of deletions. AZFa and AZFb are essential for initiating spermatogenesis, while AZFc is necessary for its completion. The AZFc in most infertile men lead to severe oligospermia (<5 X 106/mL) to azoospermia [24,25]. It spans 4.5Mb and codes for 2% candidate genes and in families of transcription units expressed in testis [26,27]. Thus, it has DAZ, basic protein Y2, Chromo Domain on Y (CDY1), Golgi autoantigen, golgin subfamily a 2 likely chondroitin sulfate proteoglycan 4 like Y, Testes Specific Transcript Y (TTTY)-linked-3, TTTY4 and TTTY17 [28]. Non-Allelic Homologous Recombination (NAHR) occurs between amplicons which include deletion, duplication and both leading to Copy Number Variation (CNV) in this sub region [29]. Hence, this region is more susceptible for microdeletions. Four partial deletions are identified viz., b2/b4, gr/gr, b2/b3 and b1/b3 having about 1.6 Mb [29-31]. Amongst these, gr/gr is the common deletion occurring due to recombination [32]. The b1/b3 differs from others, as it also encompasses the part of AZFb and results in loss of RBMY1 and PRY leading to loss of spermatogenesis in high frequency [27]. However, these partial microdeletions encompassing AZFc and AZFb regions are important in causing spermatogenic alterations in male fertility; their importance needs to be answered further. AZF deletions and semen phenotypes correlate with Yq microdeletions so, it is appropriate to consider Y deletions as a cause of testicular semen pathologies. Hence, about 25-55% of males with gonadal pathologies are related to hypospermatogenesis, arrest of sperm maturation and SCOS. Amongst, about 5-25% males are with severe oligospermia to azoospermia [32,33] harboring YCMs. But depending upon the deletion on AZF locus deleted, the phenotypic alteration varies [1,16,34]. Accordingly, the patients need to be suggested for ARTs like Intracytoplasmic Sperm Injection (ICSI) and Testicular Sperm Ejaculates (TESE).
Mechanisms of AZF Deletions
AZFa deletions are due to homologus intrachromosomal recombinations between two Human Endogenous Retroviral (HERV) sequences. AZFb, c deletions result recombination between palindromes (P5, P1 and P3, P1), respectively. Peculiar structural organisation of AZFc makes it more prone to structural rearrangements. Partial deletions occur as a result of recombination between subamplicons in AZFc. CNV occurs due to various mechanisms like gene dosage, mutation, deletion or duplication in AZF locus [1,29,35].
Yq Microdeletions Analysis
Y chromosome deletions are dynamic in addition to other genetic factors. These microdeletions involve deletions, duplications and both. Clinically, the Y chromosome changes can be categorised AZF deletions, partial AZFc deletions and the gene CNVs second to chromosomal anamolies.
Materials and Methods
Genomic DNA was used for analysing micro deletions with Sequence Tagged Sites (STS) based on Polymerase Chain Reaction (PCR) technologies. Thus, these AZF deletions were described by adopting specific markers of EAA and non EAA STS as suggested by Sen S et al., [36]. About 18 STS were essentially used for proper identification of deletions in AZF locus of infertile cases. These include SY746, SY86, DFFRY, (AZFa), SY113, SY118, SY127, RBM1Y, XKRY, SY134, SY143 (AZFb), SY153, SY148, SY157, SY255, SY254, SY158, SY160 (AZFc) in addition to SY14 (SRY). Genomic DNA was used for PCR assay and was used with necessary reagents for detection of microdeletions in AZFa, AZFb, AZFc, AZFa+b and AZFb+c sub regions and others [9,15,36,37] for ensuring optimal results [10,38-40].
Testing of Yq microdeletions has many applications for correct diagnosis for the cause of infertility [38,39]. For correlation of testicular phenotypic manifestations specific guidelines are strictly followed, while performing spermeogram analysis [41]. Accordingly, the cases were grouped into azoospermia (obstructive and non-obstructive) oligozoosospermia (5-15×106/mL), severe oligospermia, normozoospermia (normal values of sperms in the ejaculate), astherozoospermia (low level of motility, <50%), teratozoospermia (<30% of normal sperm morphology) and aspermia. Other causes include cryptorchidism, varicocele, endocrinological, obstruction of seminal pathways, infection, alcohol and chemotherapy in addition to genetic defects like cytogenetic disorders, gene mutation, Yq microdeletion [1,36,42]. Among genetic defects authors revealed the current knowledge of Yq microdeletions in the genes leading to infertility and their correlation to the phenotypic manifestations, implications, their distribution and prevalence in India.
Data Collection
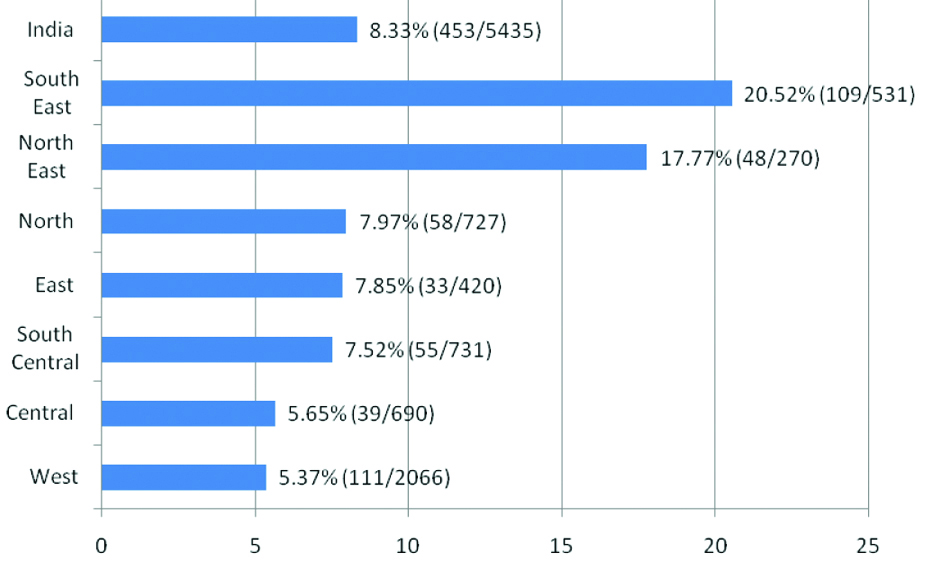
The information, latest was collected using Google on YCMs and male infertility in India and research paper collected. As cited earlier this information was categorised for the prevalence of Yq microdeletion, region wise, phenotype of semen, type of q deletions, STS markers by PCR-STS method. The data of Yq deletions were subjected to percent values if necessary, from chosen 30 study groups of seven Indian Zones; Western (Gujarat, Maharashtra); Central Zone (Lucknow, Varanasi); North (Delhi); North-East (Assam); East (West Bengal); South Central (Hyderabad) and South-East (Tamil Nadu, Kerala), respectively [Table/Fig-2]. No data was duplicated in this study.
Geographic distribution of Yq microdeletions in Indian infertile man.
Statistics
The data of 30 group studies were added according to our need with simple statistics like percentage in this cohort. No duplicate of data was observed in present study, as it might lead to erroneous information.
Results and Discussion
Zonal/Regional Deletion Analysis
In Gujarat Western part of India, screening of Yq microdeletion [43], reported 141 cases with oligospermia (41) and azoospermia (100) screen microdeletions. These authors in Anand (Gujarat) detected 24.11% (34/141) deletion with higher rate of AZFc deletions [43]. Analysis of oligospermia (3), azospermia (3), asthenospermia (2), cryptozoospermia (3), oligoasthenospermia (OAS) (4) and infertile normo-zoospermia (26) totally (41), revealed 7.31% (3/41) microdeletions [44]. In Mumbai and Nagpur, the cases were idiopathic type following the classification of World Health Organisation (WHO) [Table/Fig-3] [41] and their screening found 3.4% consisting of 56 from 1.636 infertile men, where AZFc subregion was higher in Mumbai, Maharashtra [36]. From Nagpur, Ambulkar PS and Pande SS obtained 10.6% deletion (17/160) from an analysis of oligozoospermia and azoospermia phenotypes with a dominant of AZFc followed by others [9]. A study conducted with 88 cases by Nagvenkar P et al., consisting of (42) oligospermia and (46) severe oligospermia detected only 1.1% microdeletions (1/88) with AZFc [44]. Thus, this Western region (Gujarat, Maharashtra) constituted only 5.37% (111/2066) YCM with AZFc dominance [Table/Fig-4].
Regional zonal distribution of Yq % microdeletions in Indian population.
Semen variables according to WHO [41].
| S. No. | Medical/Clinical term | Condition/Definition |
|---|
| 1 | Aspermia | Absence semen upon ejaculate |
| 2 | Zoospermia | Presence of spermatozoa in the semen |
| 3 | Azoospermia | A complete absence of sperm in the semen (both OA and NOA) |
| 4 | Normozoospermia | All normal semen values in ejaculate |
| 5 | Oligozoospermia | Presence of an abnormally low number of sperm in a semen (5-20×106/mL) |
| 6 | Severe Oligozoospermia | Sperm counts fall between 0 and 5 million sperm/mL |
| 7 | Asthenozoospermia | Reduced sperm motility |
| 8 | Oligoasthenozoospermia | Combination of reduced sperm motility and low sperm count |
| 9 | Teratozoospermia | Presence of higher abnormal morphology in the semen |
| 10 | Oligoasthenoteratozoospermia | Condition that includes low number, poor sperm movement and high abnormal forms |
| 11 | Polyzoospermia | Higher than normal values/mL |
| 12 | Hematospermia | Semen with Red Blood Cell (RBC) |
| 13 | Pyospermia | Semen with White Blood Cell (WBC) |
OA: Obstructive azoospermia; NOA: Non-obstructive azoospermia
In South Eastern (SE) region (Tamil Nadu, Kerala) six groups studied microdeletions. Abhilash VG et al., from Chennai had screened 89 cases with 34 azoospermia and 55 oligozoospermia cases for microdeletions [45]. They reported an average percent of 24.7% (22/89) consisting high frequency of AZFc. A study from Tamil Nadu, reported 72 semen samples of oligozoospermia (5), asthermo (7) and Oligoasthenozoospermia (OAS) (7), exhibited 12.9% deletions (19/72) having AZFc deletions at higher rate [46] (Sakthivel PJ and Swaminathan M, 2008). Viswambharan N, et al., documented 13.3% microdeletions (4/30) from their study cohort [47], Suganthi R et al., [48] from South Eastern Part of India described 36% deletions (18/50) in oligo and azoospermia infertile cases with higher frequency of AZFc sub-locus. Previous studies by this group [49,50] in 215 and 75 cases documented 11.2% (24/215) and 29.3% (22/75) Yq microdeletions, respectively. Thus, this zone possessed 20.52% YCMs (109/531) [Table/Fig-3].
In South Central Zone of India (Hyderabad), the results depicted in a different pattern. The screening of 20 cases of 10 each of azoospermia and oligospermic men, the observed deletions in 3 cases was 15% (3/20), with AZFc dominance [51]. Another study [52] identified in 251 infertile men only 3.98% Yq microdeletions (10/251) with high percent of AZFc deletions. There were cases of 194 idiopathic infertily and 57 were of varicocele having zoospermia followed by oligozoospermia, Oligoasthenoteratozoospermia and teratozoospermia [50]. Swarna M et al., from Hyderabad mapped 4 cases from 50 infertile cases (4/50; 8%) and 9 cases with Yq deletions (9/70, 12.8%) containing 70 idiopathic infertile cases of various testicular phenotypic manifestations with high AZFc deletions and similarly [53,54], Thangaraj K et al., from South India (Hyderabad) also reported an analysis of 340 azoospermia, where 29 had microdeletion (8.5%) of AZFc >AZFb >AZFa [55]. Thus, South Central region had the frequency of 7.52% (55/731) deletions comparatively lower than South East region [Table/Fig-3].
In Eastern region (West Bengal), recent report indicated 5% microdeletions in a study cohort documented by Ray A et al., in 80 infertile men with 19 azoospermia and 61 oligozoospermia (4/80) where AZFc was dominated [56]. Sen S et al., reported in their article, the deletions marked were 8.5% (29/340) [36]. The total average of Yq in the region remained 7.85% (33/420) [Table/Fig-3]. In North East (NE), the results varied where Mahanta R et al., [57] screened only 100 infertile cases with 5% YCM (5/100) where AZFc was dominant. In a study of Barbhuiya PN et al., published 25.30% Yq deletions in Assam (43/170) [58]. These 170 infertile men had various types of semen. They reported high frequency of AZFa and c in inferstitial deletion of AZF locus. The total deletions were of 17.17% (48/270) [Table/Fig-3].
In Lucknow and Varanasi (Central Indian Zone), various reports are available contributing 5.65% YCMs (39/690) from various groups [Table/Fig-3]. Ambasudhan R et al., detected 5% microdeletions from 177 infertile men with various phenotype manifestations in Varanasi (9/177) with high frequency of c deletions [59]. Khan FH et al., in their cohort, 100 infertile cases consisting of azoo, oligo and asthenozoospermia had 10% Yq deletions (10/100) [60]. Mainly, they found AZFa and AZFc including b+c types. The study reported by Pandey LK et al., had 64 infertile men with 3.3% deletions (2/64), where AZFc was dominant in their semen types [61]. Similarly, Singh K and Raman R in their study obtained 4.8% (13/270) in azoo- and oligospermic men with infertility possessing of AZFc and AZFd deletions [62]. In Lucknow Mittal RD et al., in 79 infertile men, with oligo (25), azoospermia (54), screened for Y Chromosome Microdeletion (YCM), who reported 6.5%(5/79) with AZFc and AZFb deletions followed by b+c types [63].
In Northern Part of India (Delhi), Dada R et al., documented 6% deletions in a study having oligo and azoospermia (8/133) [64]. Earlier study by these researchers in 70 infertile cases with varicocele, 11.4% microdeletions was reported (8/70) [65]. Mitra A et al., in their study group containing 14 azoospermia with Klinefelter Syndrome (KFS) had 4 cases of AZFa and AZFc deletion (4/14; 28.6%) [66]. In another study with 119 azoo and 51 oligospermia cases had 5.29% microdeletions (9/170), spanning AZFc followed by b+c a and b deletions [67]. Recently, Dada R et al., analysed 140 cases of oligo-and azoo with varicocele and found 5.7% deletions (8/140) [68]. They observed AZFc and a+b are higher than b and b+c deletions. Sachdeva K et al., claimed that out of 200 infertile cases with spermatogenic failure at testicular level had deletions 10.5% (21/200) [69]. The protocol for identification of Y deletions in their cases was of Non Obstructive Azoospermic (NOA) and oligospermic men. Hence, in north zone, the average YCMs are 7.97% (58/727) having dominance of AZFc subregion [Table/Fig-3].
This mapping of Yq microdeletions in Indian studies of these 30 groups/citations indicated a range from 1.1 to 28.6% with an average frequency of 8.33% from total analysis of 5435 chromosomes (453/5435). It includes different groups, region and diverse classes of infertile men. Sub continent analysis of large databases has earlier revealed the percent of microdeletions ranged from 8-10% [70], in support of the observations. But reports presented by Sen S et al., had significantly lower frequencies (3.4%, 56/1636; 5.8% 215/3647) than the data (8.33%) [Table/Fig-3] [36].
AZF Locus Microdeletions
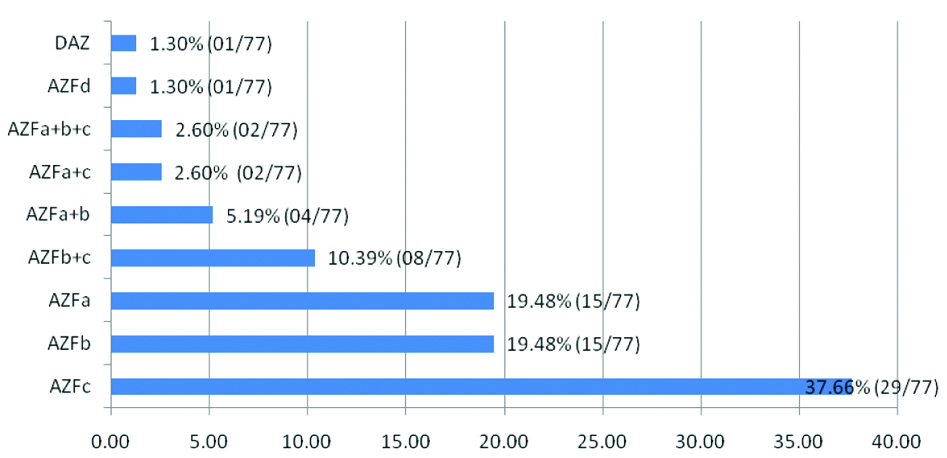
AZF microdeletions and semen phenotypic correlations are well appreciated in relation to infertility by several authors [1,17,37,71]. Overall 25-55% males with testicular pathologies like hypo spermatogenesis, sperm maturation arrest, Sertoli Cell Only Syndrome (SCOS) and about 5-25% males are affected by SOS or azoospermia harbouring YCMs as indicated earlier [32]. In this cohort more azoospermia cases (referred/selected 66%; affected 61%) in 30 and 15 study groups, respectively, than oligo-and others are attributed to AZFc locus [Table/Fig-2,3]. The sub-region microdeletions delivered in present study were AZFc (37.66%; 29/77), AZFa and AZFb (19.48%; each15/77), AZFb+c (10.39%; 8/77), AZFa+b (5.19%; 5/77), AZF a+c and AZFa+b+c (2.6%;each 2/77) and 1.3% each of AZFd and DAZ (1/7) respectively in a total of 77 deletions in present study [Table/Fig-5].
Summary of percent (%) AZF sub region micro deletions (77) in our study.
AZFa Microdeletions and Phenotype Correlation
Microdeletions in this region contributed to 5%, as it contains only 4 single copies of genes [16,17]. Mostly these infertile cases have spermotogenic failure and restricted to SCOS [33,72,73]. Few workers believe it is related to oligospermia [33]. In present study, few study groups (7/30) mapped high rate of AZFa deletion and related to oligo/severe oligospermia. Thus, this region deletion depends on amount of genetic material deleted ranging from azoospermia to normozoospermia [74,75]. Present study review data thus had indicated low frequency of microdeletion contributing to testicular pathology [Table/Fig-6] as seven group had AZFa deletions at higher rate.
Summary of referred/screened phenotypes cases vs AZF locus with percent deletion in Indian population. @ Sr. No. 2 and 17: Same but Indian Zones are compared from this citation.
| Sr. No. | Author | Phenotypes selected | AZF Locus detected | Deleted/Total cases | Percent deletion (%) | Locations | Indian zones |
|---|
| Azoo | Oligo/SOS | Others |
|---|
| 1 | Nailwal M and Chauhan JB [43] | 41 | 100 | - | AZFc, AZFb, AZFa | 34/141 | 24 | Anand | Western |
| 2 | Sen S et al., [36] | 992 | 600 | 44 | AZFc, b+c, a, b, a+b, a+c, a+b+c | 56/1636 | 3.4 | Mumbai |
| 3 | Shah PS et al., [42] | 3 | 3 | 35 | AZFc, AZFa | 3/41 | 7.3 | Ahmedabad |
| 4 | Ambulkar PS and Pande SS [9] | 90 | 70 | - | AZFc, b+c, b, a, a+b | 17/160 | 10.6 | Nagpur |
| 5 | Nagvenkar P et al., [44] | 40 | 48 | - | AZFc | 1/88 | 1.1 | Mumbai |
| 6 | Babu SR et al., [51] | 10 | 10 | - | AZFc | 3/20 | 15 | Hyderabad | South central |
| 7 | Rao L et al., [52] | 104 | 90 | 57 | AZFc, a=b+ | 10/251 | 3.89 | Hyderabad |
| 8 | Swarna M et al., [54] | 15 | 55 | - | AZFc | 9/70 | 12.8 | Hyderabad |
| 9 | Swarna M et al., [53] | 10 | 40 | - | AZFc | 4/50 | 8 | Hyderabad |
| 10 | Thangaraj K et al., [55] | 340 | - | - | AZFc, b, a | 29/340 | 8.5 | Hyderabad |
| 11 | Suganthi R et al., [48] | 30 | 20 | - | AZFc, b+c, a=b+ | 18/50 | 36 | Tamil Nadu | South east |
| 12 | Suganthi R et al., [49] | 215 infertile cases | AZFc, b,b+c, a+b+c, a+c, a | 24/215 | 11.2 | Tamil Nadu |
| 13 | Suganthi R et al., [50] | 75 infertile cases | - | 22/75 | 29.3 | Tamil Nadu |
| 14 | Vishwambaran N et al., [47] | 17 | 13 | - | AZFc, a=b+ | 4/30 | 13.3 | Tamil Nadu |
| 15 | Sakthivel PJ and Swaminathan M [46] | 72 infertile cases | AZFc, a | 19/72 | | | 12.9 | Coimbatore |
| 16 | Abhilash VG et al. [45] | 34 | 54 | - | AZFc | 22/89 | 24.7 | Chennai |
| 17 | Sen S et al., [36] | 340 infertile cases | - | 29/340 | 8.5 | West Bengal | Eastern |
| 18 | Ray A et al., [56] | 19 | 6 | - | AZFc | 4/80 | 5 | West Bengal |
| 19 | Mahanta R et al., [57] | 100 infertile cases | AZFc | 5/100 | 5 | Assam | North east |
| 20 | Barbhuiya PN et al., [58] | 50 | 82 | 38 | AZFc, d | 43/170 | 25.3 | Assam |
| 21 | Khan FH et al., [60] | 100 infertile cases | AZFa, c, b+c | 10/100 | 10.0 | Varanasi | Central |
| 22 | Pandey LK et al., [61] | 64 infertile cases | AZFc | 2/64 | 3.48 | Varanasi |
| 23 | Singh K and Raman R [62] | 270 infertile cases | AZFc, DAZ, AZFb | 13/270 | 4.80 | Varanasi |
| 24 | Mittal RD et al., [63] | 54 | 24 | - | AZFc, b | 5/79 | 6.3 | Lucknow |
| 25 | Ambasudhan R et al., [59] | 142 | 33 | 2 | AZFc | 9/177 | 5.1 | Varanasi |
| 26 | Mitra A et al., [66] | 14 infertile cases | AZFa, c | 4/14 | 28.6 | New Delhi | North zone |
| 27 | Mitra A et al., [67] | 119 | 51 | - | AZFc, a=b+, b+c | 9/170 | 5.3 | New Delhi |
| 28 | Dada R et al., [65] | 40 | 30 | - | AZFc, b, a+b | 8/70 | 11.4 | New Delhi |
| 29 | Dada R et al., [64] | 133 infertile cases | AZFc, b, a, a+b | 8/133 | 6 | New Delhi |
| 30 | Dada R et al., [68] | 114 | 16 | - | AZFc, a+b, b, b+c | 8/140 | 5.7 | New Delhi |
| 31 | Sachdeva K et al., [69] | 200 infertile cases | AZFc, a,b, b+c | 21/200 | 10.5 | New Delhi |
| Total (30)@ | | Deletion types | 453/5435 | 8.33% | | All Zones (7) |
Deletion frequencies in study groups (30):AZFc*(27)>AZFb**(9)>AZFa***(7). Numbers in parenthesis indicate study groups in the [Table/Fig-3] (1-30); *:1-16, 18, 19, 20, 22-25, and 27-31 (27 study groups)
**:1, 4, 10, 12, 23, 24, 28-30 (9 Study groups); ***: 2, 3, 15, 21, 26, 31 (6 Study groups); +=equal.: Total deletion=77; Referred/Selected cases (21 Groups); Azoospermia=66%(2264/3445);OS/SOS=30% (1045/3445); Others=4%(136/3445); ΨSOS: Severeoligozoospermia
AZFb Deletion and Phenotypes
This region is also comparatively smaller than c region containing 3.2 Mb spanning few genes [76] that are single and multiple copies of DNA repeats and 14 amplicons and contributes about 16% deletions. This region is important for early stages of spermatogenesis and frequently responsible for hyperspermatogenesis [77]. In present review report, AZFb microdeletions exhibited high frequency of these deletions found relatively in 9 study groups (9/30) [Table/Fig-6]. Phenotypes range from spermatogenic anomalies and oligo- spermia/hypo-oligospermia [42,76]. The deletions hence in this AZFb, however had more than AZFa types in infertility cases with oligo to SOS [Table/Fig-6] [11,36].
AZFc Deletions and Phenotype Correlation
The AZFc region comprises of repeated sequences and palindromes making it suitable for deletions with partial types. Hence, these deletions correlated with testicular phenotypes ranging from severe oligospermia to azoospermia [27,78,79]. Patients having these deletions show progressive decline in sperm counts from oligo to severe and absence of sperm [80-83]. In our cited study groups (30) [Table/Fig-6], more (66%) infertile men were referred for screening and affected (61%) were azoospermia followed by oligospermia correlating with high frequency of AZFc deletions comparatively. Colaco S and Modi D; Sen S et al., and Shah PS et al., and several researchers also noticed high prevalence of AZFc microdeletions due to its complex structure [1,36,42]. Twenty seven study groups (27/30) delivered high rate of AZFc microdeletions amongst all 30 groups [Table/Fig-6].
Other Microdeletions and Phenotypes
Few study groups in our cohort reported AZFb+c, AZFa+b, AZFa+c, AZFa+b+c, AZFd and DAZ having other double and triple deletions that are related to hypospermetogenesis and other testicular pathologies. These deletions were low in frequency as compared to AZFc, a and b types and are depicted in [Table/Fig-6]. Ambulkar PS and Pande SS and Suganthi R et al., detected high rate of AZFc microdeletions comparing to abnormal semen types in comparison to a and b deletions [9,37]. Dada R et al., in their studies revealed high rate of AZF a+b over AZFb micro deletions [Table/Fig-6] [65,68].
Partial Microdeletions and Phenotypes
The AZFc has partial deletions viz b2/b4, b1/b3, b2/b3 and gr/gr, where b1/b3 combination of AZFb and AZFc regions in addition to CNV. Among, gr/qr the most common type that are due to homologous recombination. These partial microdeletions are highly variable in multiple studies reported and are controvertial [1,28]. Similarly few reports are documented regarding to CNVs [1]. In present report, these are not well screened by any group and require further elucidation.
Prevalence and Distribution of Yq Microdeletions in India
Overall distribution of microdeletion screening notified that AZFc region has high frequency as it is a complexed structure. Most of the testicular phenotype cases with azoospermia followed by oligospermia possessed deletion of AZFc only. It is overall followed by AZF region of a, b, b+c and others [Table/Fig-6] to support the data of previous authors [36,37] in Indian infertile men.
In our review cohort, further South East (20.52%), North East (17.77%), North (7.97%), East (7.85%) and South Central (7.52%) contained comparatively high frequency of Yq deletions followed by Western (5.37%) and Central (5.65%) regions, respectively. Thus these YCM are minimum in Western and Central Indian Zones [Table/Fig-2,3]. This could be due to geographic regions, ethnicity, sample size, population, food styles, STS-kits used and other epigenetic factors. Such results with other authors are also documented in Indian population in related to prevalence and distribution of Yq microdeletions [36,42].
Implications
Yq Microdeletions Analysis and Methodologies in Future
It is suggested that analysis of Yq microdeletions involve use of multiple STS markers spanning various AZF loci [37]. Authors used EAA and non-EAA markers of 4 to 30, but it is better to use a good number of markers to detect Yq microdeletions. Further, EAA and EMQN strongly recommend two, STS which is specific to DAZ gene in the P2 and P1 palindromes. Partial deletions cited to b2/b4 pattern can also be analysed using Sy 160 STS markers. Conventional PCR is to be upgraded to multiplex PCR, which is less costlier less time and advantageous than former [23]. Commercial kits such as Diachem/Bird, Euroclone are available [84]. Bunyan DJ et al., reported a method for the detection of partial AZFc deletions and Multiplex Ligation dependant Probe Amplification (MLPA) probe mix (P360) known as MLPA assay [85]. Microarray developed by Osborne EC et al., for microdeletions are also recommended for these studies with better reproducible results to infertile men [86].
Microdeletions and ARTs
Mapping of microdeletions of Yq in infertile men correlates to phenotype of testis which is a good predictor of sperm retrieval during Testicular Sperm Ejaculates (TESE). Microdeletions in infertile cases carry them to offspring born after ICSI. Further, testing of sperm for microdeletions, which are useful for sperm banks related to ARTs. Sperm carrying higher microdeletions may lead poor quality embryos [87] after using ICSI. Further, patients with AZFc microdeletions presented high sperm recovery from testis than cases with AZFa and AZFb deletions who presented a poor prognosis [88]. Simoni M et al.,; Nailwal M and Chauhan JB also supported azoospermic cases with AZFc deletions are better for ICSIs [15,75]. Genetic counselling is adapted to couples undergoing such ARTs. Moreover, prediction of prognosis of male infertility with Yq microdeletion is also important for recovery of testicular phenotypes. Testing of AZF region for deletions/CNVs may also be essential for detecting testicular tumours. Additionally, microdeletions causing infertility in males undergoing ARTs through genetic counselling are also strongly associated with neuropsychiatric disorders [1].
Y-Deletion and DNA Damage
DNA fragmentation increases with Oxidative Stress (OS) in a sperm cell. This OS occurs in sperm of infertile cases by several factors such as heavy metals, free radicals, caspases during apoptosis. Such induced OS upsets oxido-redox ratios leading to DNA damage and mutations. Hence, it is proved that DNA Fragmentation Index (DFI) shoots up in sperm of infertile men due to reduced recombination repair, DNA package anomaly [17,89]. Thus, OS is implicated for DNA damage inturn relating to Yq microdeletions in infertile cases. Hence, OS DNA damage and deletion in Y chromosome are related in infertile males [70,90-93], but such couples with affected males need to be treated with antioxidants and evaluated and such cases are also to be counselled prior to adopt ICSI and other IVF techniques in future [34,37,42,94,95]. [Table/Fig-7] shows semen types deleted vs deletion correlation in 15 groups of Indian population. Azoospermia cases (163/266=61%) were affected than others types.
Semen types deleted vs deletion correlation in 15 groups of Indian population. Azoospermia cases (163/266=61%) were affected than others.
| Sr. No | Author | Semen types deleted | Deleted/Total |
|---|
| Azoo | oligo/SOS | Others | |
|---|
| 1 | Sen S et al., [36] | 34 | 22 | - | 56/1636 (3.4%) |
| 2 | Shah PS et al., [42] | 1 | 2 | - | 3/41 (7.3%) |
| 3 | Suganthi R et al., [48] | 10 | 8 | - | 18/50 (36%) |
| 4 | Barbhuiya PN et al., [58] | 13 | 18 | 12 | 43/170 (28.3%) |
| 5 | Ambasudhan R et al., [59] | 8 | - | 1 | 9/177 (5%) |
| 6 | Nagrvenkar P et al., [44] | 1 | - | - | 1/88 (1.1%) |
| 7 | Nailwal M and Chauhan JB [43] | 20 | 14 | - | 34/141 (24.11%) |
| 8 | Rao L et al., [52] | 5 | 4 | 1 | 10/25 (3.89%) |
| 9 | Ambulkar PS and Pande SS [9] | 10 | 7 | - | 17/160 (10.6%) |
| 10 | Thangaraj K et al., [55] | 34 | - | - | 34/25 (8.5%) |
| 11 | Swarna M et al., [53] | 2 | 1 | 1 | 4/80 (8%) |
| 12 | Swarna M et al.. [54] | 4 | 1 | 4 | 9/70 (12.8%) |
| 13 | Dada R et al., [64] | 7 | 1 | - | 8/133 (6.0%) |
| 14 | Dada R et al., [68] | 6 | 2 | - | 8/140 (5.7%) |
| 15 | Athalye AS et al., [94] | 8 | 3 | 1 | 12/100 (12%) |
| Total (15 Groups) | 163 | 83 | 20 | 266/3036 (8.76%) |
YCMs in Indian Scenario
Published data earlier showed the frequency of microdeletions in Indian population ranged from 3 to 29.34% with an average frequency of 8.1% [37,42,47,61,65]. Thangaraj K et al., proposed 8.5% YCMs from the study of 340 azoospermia infertile cases [55]. In another study, it is reported that it increased to 9.63% [65]. Only Sen S et al., [36] documented to 3.4% frequency of deletions in Indian population of 1636 cases, which is significantly lower than others. Same group had also mentioned low frequency 5.8% deletions, but present study cohort exhibited 8.33% deletions from 5435 Y chromosomes analysed supporting the data of Thangaraj K et al., Pandey LK et al., and Dada R et al., [55,61,64]. Recently, Shah PS et al., and Rao MV, also reported similar data falling in range of 8-10% [16,42] and Waseem AS et al., in India these deletions ranged from 0.59-32.62% with an average of 13.48% [95], but his own data delivered only 10.02% deletion frequency. This variation could be due to ethnic background study protocol and other factors. The Indian population further is affected by AZFc deletion followed by AZFb and AZFa in most of the testicular phenotypes as reported by others [37]. The azoospermia cases thus considered to be mostly affected than oligo/severe oligospermia and others. The AZFc deletions containing such azoospermic cases are better suitable than other pathologic phenotypes as suggested above [75,88,95] for associated reproductive technologies in present and future of Indian Scenario.
Limitation(s)
Male infertility caused by genetic factor needs to be assessed with proper genetic tests. Significance of microdeletions/partial deletions are to be communicated to couples. Accurate methodology is to be developed for full success rate of deletions. Congenital malformations, genetic defects, loss of birth weight and its propagation to offspring are to be limited successfully.
Conclusion(s)
This study cohort clearly enumerates the Yq microdeletion (8.33%) leading to male infertility mainly falling in the range of 8-10% in India. Correlation between these types of deletions and associated testicular phenotypes provide identification of infertility type, detecting the prognosis of infertile males, treatment and other defects like cancer. Azoospermic men are affected with high frequency of AZFc deletions in present study having more percent in Eastern region of India. Herewith, it also provides opportunities for counseling the couple adopting ARTs, where male partner carries these specific deletions. Y chromosome deletion analysis is also necessary in sperm banks to curtile multiple abnormal embryo yield after ICSI and TESI, since the prevalence and distribution, of these complete and partial deletions vary region wise, population groups, ethnicity, sample size, STS marker used, selection criteria, haplotypes, environment and other epigenetic factors.
Further, hereby these studies definitely take forward new direction to evaluate infertile males possessing Yq deletions for correct treatment clinically. These advancements allow more widespread of this deletion screening and its implication in infertility clinics, IVF and andrology laboratories in our country and also around the globe in future.
[1]. Colaco S, Modi D, Genetics of the human Y chromosome and its association with male infertilityReprod Biol Endocrinol 2018 16(1):1410.1186/s12958-018-0330-529454353 [Google Scholar] [CrossRef] [PubMed]
[2]. Vogt PH, Bender U, Zimmer J, Strowitzki T, Human Y chromosome and male infertility: Forward and back from azoospermia factor chromatin structure to azoospermia factor gene functionMonographs in Human Genetics 2017 21:57-73.10.1159/000477278 [Google Scholar] [CrossRef]
[3]. Manz E, Alkan M, Bühler E, Schmidtke J, Arrangement of DYZ1 and DYZ2 repeats on the human Y-chromosome: A case with presence of DYZ1 and absence of DYZ2Mol Cell Probes 1992 6(3):257-59.10.1016/0890-8508(92)90025-S [Google Scholar] [CrossRef]
[4]. Esteves SC, Agarwal A, Novel concepts in male infertilityItl Braz Jurl 2011 37(1):05-15.10.1590/S1677-5538201100010000221385475 [Google Scholar] [CrossRef] [PubMed]
[5]. Colaco S, Modi D, Consequences of Y chromosome microdeletions beyond male infertilityJournal of Assisted Reproduction and Genetics 2019 36(1):1329-37.10.1007/s10815-019-01492-z3121488 [Google Scholar] [CrossRef] [PubMed]
[6]. Lahn BT, Page DC, Functional coherence of the human Y chromosomeScience 1997 278:675-80.10.1126/science.278.5338.6759381176 [Google Scholar] [CrossRef] [PubMed]
[7]. Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Mammalian Y chromosomes retain widely expressed dosage-sensitive regulatorsNature 2014 508(7497):494-99.10.1038/nature1320624759411 [Google Scholar] [CrossRef] [PubMed]
[8]. Tiepolo L, Zuffardi O, Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long armHuman Genetics 1976 34(2):119-24.10.1007/BF002788791002136 [Google Scholar] [CrossRef] [PubMed]
[9]. Ambulkar PS, Pande SS, Study of Y-Chromosome microdeletions in azoospermic infertile males using multiplex PCR analysisBiosciences Biotechnology Research Asia 2018 15(2):351-57.10.13005/bbra/2639 [Google Scholar] [CrossRef]
[10]. Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, Molecular and clinical characterization of Y chromosome microdeletions in infertile men: A 10-year experience in ItalyJ Clin Endocrinol Metab 2007 92(3):762-70.10.1210/jc.2006-198117213277 [Google Scholar] [CrossRef] [PubMed]
[11]. Krausz C, McElreavey K, Y chromosome microdeletions infertile’malesHuman Reproduction 2001 16(6):130610.1093/humrep/16.6.130611387311 [Google Scholar] [CrossRef] [PubMed]
[12]. Vogt P, Chandley AC, Hargreave TB, Keil R, Ma K, Sharkey A, Microdeletions in interval 6 of the Y chromosome of males with idiopathic sterility point to disruption of AZF, a human spermatogenesis geneHuman Genetics 1992 89(5):491-96.10.1007/BF002191721634226 [Google Scholar] [CrossRef] [PubMed]
[13]. Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein geneNat Genet 1995 10:383-93.10.1038/ng0895-3837670487 [Google Scholar] [CrossRef] [PubMed]
[14]. Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11Human Molecular Genetics 1996 5(7):933-43.10.1093/hmg/5.7.9338817327 [Google Scholar] [CrossRef] [PubMed]
[15]. Simoni M, Bakker E, Krausz C, EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletionsState of the art 2004. Int J Androl 2004 27:240-49.10.1111/j.1365-2605.2004.00495.x15271204 [Google Scholar] [CrossRef] [PubMed]
[16]. Rao MV, Y chromosome microdeletion and its association with male infertility. 2019. 5th Proc. A. P. Science Congress Abstr. PP XXII. 28-30, NovSrikakulam [Google Scholar]
[17]. Cotter PD, Norton ME, Y chromosome heterochromatin variation detected at prenatal diagnosisPrenat Diagn 2005 25(11):1062-63.10.1002/pd.128016299832 [Google Scholar] [CrossRef] [PubMed]
[18]. Vogt PH, Fernandes S, Polymorphic DAZ gene family in polymorphic structure of AZFc locus: Artwork or functional for human spermatogenesis? Review articleAPMIS 2003 111(1):115-27.10.1034/j.1600-0463.2003.11101161.x12752250 [Google Scholar] [CrossRef] [PubMed]
[19]. Vogt PH, Human chromosome deletions in Yq11, AZF candidate genes and male infertility: History and updateMHR: Basic Science of Reproductive Medicine 1998 4(8)10.1093/molehr/4.8.7399733430 [Google Scholar] [CrossRef] [PubMed]
[20]. Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I, The choice and outcome of the fertility treatment of 38 couples in whom the male partner has a Yq microdeletionHum Reprod 2005 20:1887-96.10.1093/humrep/deh84715790609 [Google Scholar] [CrossRef] [PubMed]
[21]. Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, Gianotten J, A family of human Y chromosomes has dispersed throughout northern eurasia despite a 1.8-Mb deletion in the azoospermia factor c regionGenomics 2004 83(6):1046-52.10.1016/j.ygeno.2003.12.01815177557 [Google Scholar] [CrossRef] [PubMed]
[22]. O’Flynn O’Brien KL, Varghese AC, Agarwal A, The genetic causes of male factor infertility: A reviewFertil Steril 2010 93:01-12.10.1016/j.fertnstert.2009.10.04520103481 [Google Scholar] [CrossRef] [PubMed]
[23]. Alechine E, Corach D, High-throughput screening for spermatogenesis candidate genes in the AZFc region of the Y chromosome by multiplex real time PCR followed by high resolution melting analysisPLoS ONE 2014 9(5):e9722710.1371/journal.pone.009722724828879 [Google Scholar] [CrossRef] [PubMed]
[24]. Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, The male-specific region of the human Y chromosome is a mosaic of discrete sequence classesNature 2003 423(6942):825-37.10.1038/nature0172212815422 [Google Scholar] [CrossRef] [PubMed]
[25]. Song SH, Chiba K, Ramasamy R, Lamb DJ, Recent advances in the genetics of testicular failureAsian J Androl 2016 18:350-55.10.4103/1008-682X.17885727048782 [Google Scholar] [CrossRef] [PubMed]
[26]. Yu XW, Wei ZT, Jiang YT, Zhang SL, Y chromosome azoospermia factor region microdeletions and transmission characteristics in azoospermic and severe oligozoospermic patientsInt J Clin Exp Med 2015 8(9):14634-46. [Google Scholar]
[27]. Rozen SG, Marszalek JD, Irenze K, Skaletsky H, Brown LG, Oates RD, AZFc deletions and spermatogenic failure: A population-based survey of 20,000 Y chromosomesAm J Hum Genet 2012 91(5):890-96.10.1016/j.ajhg.2012.09.00323103232 [Google Scholar] [CrossRef] [PubMed]
[28]. Nailwal M, Chauhan JB, Azoospermia factor C subregion of the Y chromosomeJ Hum Reprod Sci 2017a 10(4):256-60.10.4103/jhrs.JHRS_16_1729430151 [Google Scholar] [CrossRef] [PubMed]
[29]. Lu C, Jiang J, Zhang R, Wang Y, Xu M, Qin Y, Gene copy number alterations in the azoospermia-associated AZFc region and their effect on spermetogenic impairmentMol Hum Reprod 2014 20:836-43.10.1093/molehr/gau04324935076 [Google Scholar] [CrossRef] [PubMed]
[30]. Visser L, Westerveld GH, Korver CM, van Daalen SK, Hovingh SE, Rozen S, Y chromosome gr/gr deletions are a risk factor for low semen qualityHum Reprod 2009 24:2667-73.10.1093/humrep/dep24319602516 [Google Scholar] [CrossRef] [PubMed]
[31]. Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C, Vogt PH, A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup NAm J Hum Genet 2004 74:180-87.10.1086/38113214639527 [Google Scholar] [CrossRef] [PubMed]
[32]. Bansal SK, Jaiswal D, Gupta N, Singh K, Dada R, Sankhwar SN, Gr/gr deletions on Y-chromosome correlate with male infertility: An original study, meta-analyses, and trial sequential analysesScientific Reports 2016 6:1979810.1038/srep1979826876364 [Google Scholar] [CrossRef] [PubMed]
[33]. Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, Mielnik A, High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndromeMol Hum Reprod 2001 7:987-94.10.1093/molehr/7.10.98711574668 [Google Scholar] [CrossRef] [PubMed]
[34]. Modi D, Gene copy Number variation in AZFc locus of the human Y chromosome and its implication in male fertility 2018 Hyderabad. Abstr. I-23 [Google Scholar]
[35]. Noordam MJ, Westerveld GH, Hovingh SE, van Daalen SK, Korver CM, van der Veen F, Gene copy number reduction in the azoospermia factor c (AZFc) region and its effect on total motile sperm countHuman Molecular Genetics 2011 20(12):2457-63.10.1093/hmg/ddr11921429917 [Google Scholar] [CrossRef] [PubMed]
[36]. Sen S, Pasi AR, Dada R, Shamsi MB, Modi D, Y chromosome microdeletions in infertile men: Prevalence, phenotypes and screening markers for the Indian populationJ Assist Reprod Genet 2013 30(3):413-22.10.1007/s10815-013-9933-023344732 [Google Scholar] [CrossRef] [PubMed]
[37]. Suganthi R, Vijesh VV, Vandana N, Fathima ali benazir J, Y choromosomal microdeletion screening in the workup of male infertility and its current status in IndiaInt J Fertil Steril 2014 7(4):253-66. [Google Scholar]
[38]. Chang PL, Sauer MV, Brown S, Y chromosome microdeletion in a father and his four infertile sonsHuman Reproduction 1999 14(11):2689-94.10.1093/humrep/14.11.268910548602 [Google Scholar] [CrossRef] [PubMed]
[39]. Gatta V, Stuppia L, Calabrese G, Morizio E, Guanciali-Franchi P, Palka G, A new case of Yq microdeletion transmitted from a normal father to two infertile sonsJournal of Medical Genetics 2002 39(6):e2710.1136/jmg.39.6.e2712070259 [Google Scholar] [CrossRef] [PubMed]
[40]. Foresta C, Moro E, Ferlin A, Y chromosome microdeletions and alterations of spermatogenesisEndocr Rev 2001 22(2):226-39.10.1210/edrv.22.2.042511294825 [Google Scholar] [CrossRef] [PubMed]
[41]. WHO. Laboratory manual for the examination and processing of human semen. 5. Geneva: World Health Organization; 2010 [Google Scholar]
[42]. Shah PS, Shah NP, Shah SC, Patel T, Raval RJ, Dominic J, Y chromosome microdeletion and phenotype correlates with male infertility in Gujarati populationEJMS 2020 5(1):19-21.10.32677/EJMS.2020.v05.i01.006 [Google Scholar] [CrossRef]
[43]. Nailwal M, Chauhan JB, Gene scanning for microdeletions in the azoospermia factor region of Y-chromosome in infertile men of Gujarat, IndiaJ Clin Diagn Res 2017 11(8):GC01-GC06.10.7860/JCDR/2017/26750.1035028969154 [Google Scholar] [CrossRef] [PubMed]
[44]. Nagvenkar P, Desai K, Hinduja I, Zaveri K, Chromosomal studies in infertile men with oligozoospermia & non-obstructive azoospermiaIndian Journal of Medical Research 2005 122(1):34 [Google Scholar]
[45]. Abhilash VG, Saraswathy R, Marimuthu KM, The frequency of Y chromosome microdeletions in infertile men from Chennai, a South East Indian population and the effect of smoking, drinking alcohol and chemical exposure on their frequenciesInternational Journal of Genetics and Molecular Biology 2010 2(7):147-57. [Google Scholar]
[46]. Sakthivel PJ, Swaminathan M, Y chromosome microdeletions in sperm DNA of infertile patients from Tamil Nadu, south IndiaIndian J Urol 2008 24(4):480-85.10.4103/0970-1591.4425219468501 [Google Scholar] [CrossRef] [PubMed]
[47]. Viswambharan N, Suganthi R, Simon AM, Manonayaki S, Male infertility: Polymerase chain reaction-based deletion mapping of genes on the human chromosomeSingapore Med J 2007 48:1140-42. [Google Scholar]
[48]. Suganthi R, Vijesh V, Jayachandran S, Fathima benazir JA, Multiplex PCR based screening for microdeletions in azoospermia factor region of Y chromosome in azoospermic and severe oligozoospermic south Indian menIran J Reprod Med 2013 11(3):219-26. [Google Scholar]
[49]. Suganthi R, Manonayaki S, Benazir JF, Molecular analysis of Y-chromosome microdeletions in infertile menInt J Med Sci 2009 2:54-60. [Google Scholar]
[50]. Suganthi R, Vijesh VV, Chitra J, Sreelekha G, Ali Fathima Benazir J, Rapid detection of Y chromosome microdeletions in infertile south Indian men with oligo- or azoospermia. Proceedings of International Conference on Genomics and ProteomicsNational Institute of Technology (NIT) 2011 O216:79-84.Calicut [Google Scholar]
[51]. Babu SR, Swarna M, Padmavathi P, Reddy PP, PCR analysis of Yq microdeletions in infertile males, a study from South IndiaAsian J Androl 2002 4:265-68. [Google Scholar]
[52]. Rao L, Babu A, Kanakavalli M, Padmalatha V, Singh A, Singh PK, Chromosomal abnormalities and y chromosome microdeletions in infertile men with varicocele and idiopathic infertility of South Indian originJ Androl 2004 25:147-53.10.1002/j.1939-4640.2004.tb02770.x14662798 [Google Scholar] [CrossRef] [PubMed]
[53]. Swarna M, Babu SR, Reddy PP, AZFc deletions in idiopathic infertile males from south IndiaInternational Journal of Human Genetics 2003 3(1):01-04.10.1080/09723757.2003.11885819 [Google Scholar] [CrossRef]
[54]. Swarna M, Babu SR, Reddy PP, Y chromosome microdeletions in infertile males from Andhra Pradesh, South IndiaGenet Test 2004 8:328-35.10.1089/gte.2004.8.32815727259 [Google Scholar] [CrossRef] [PubMed]
[55]. Thangaraj K, Gupta NJ, Pavani K, Y chromosome deletions in azoospermic men in IndiaJ Androl 2003 24(4):588-97.10.1002/j.1939-4640.2003.tb02710.x12826698 [Google Scholar] [CrossRef] [PubMed]
[56]. Ray A, Tapadar A, Kar M, Kundu R, Nandy S, Microdeletions in the Y chromosome in cases of male infertility in a population of West BengalJ Anatl Soc Insia 2014 63:52-56.10.1016/j.jasi.2014.04.002 [Google Scholar] [CrossRef]
[57]. Mahanta R, Gogoi A, Roy S, Bhattacharyya IK, Sharma P, Prevalence of azoospermia factor (AZF) deletions in idiopathic iinfertile males in North-East IndiaInt J Hum Genet 2011 11:99-104.10.1080/09723757.2011.11886130 [Google Scholar] [CrossRef]
[58]. Barbhuiya PN, Gogoi A, Goenka D, Ahmed GU, Mahanta R, Specific genetic marker based molecular study of the azfa & azfd region microdeletion in infertile cases of northeast IndiaInt J Biol Med Res 2013 4:3121-27. [Google Scholar]
[59]. Ambasudhan R, Singh K, Agarwal JK, Singh SK, Khanna A, Sah RK, Idiopathic cases of male infertility from a region in India show low incidence of Y- chromosome microdeletionJ Biosci 2003 28:605-12.10.1007/BF0270333614517364 [Google Scholar] [CrossRef] [PubMed]
[60]. Khan FH, Ganesan P, Kumar S, Y chromosome microdeletion and altered sperm quality in human males with high concerntration of seminal heaxchlorocyclohexane (HCH) Chemosphere 2010 80:972-77.10.1016/j.chemosphere.2010.05.04720561669 [Google Scholar] [CrossRef] [PubMed]
[61]. Pandey LK, Pandey S, Gupta J, Saxena AK, Loss of the AZFc region due to a human Y-chromosome microdeletion in infertile male patientsGenet Mol Res 2010 9:1267-73.10.4238/vol9-2gmr83620603812 [Google Scholar] [CrossRef] [PubMed]
[62]. Singh K, Raman R, Male infertility: Y-chromosome deletion and testicular aetiology in cases of azoo-/oligospermiaIndian J Exp Biol 2005 43:1088-92. [Google Scholar]
[63]. Mittal RD, Singh G, Srivastava A, Pradhan M, Kesari A, Makker A, Y chromosome micro-deletions in idiopathic infertility from Northern IndiaAnn Genet 2004 47:331-37.10.1016/j.anngen.2004.05.00315581830 [Google Scholar] [CrossRef] [PubMed]
[64]. Dada R, Gupta NP, Kucheria K, Yq microdeletions-azoospermia factor candidate genes and spermatogenic arrestJ Biomol Tech 2004 15:176-83. [Google Scholar]
[65]. Dada R, Gupta NP, Kucheria K, AZF microdeletions associated with idiopathic and non-idiopathic cases with cryptorchidism and varicoceleAsian J Androl 2002 4:259-63. [Google Scholar]
[66]. Mitra A, Dada R, Kumar R, Gupta NP, Kucheria K, Gupta SK, Y chromosome microdeletions in azoospermic patients with Klinefelter’s syndromeAsian Journal of Andrology 2006 8(1):81-88.10.1111/j.1745-7262.2006.00083.x16372123 [Google Scholar] [CrossRef] [PubMed]
[67]. Mitra A, Dada R, Kumar R, Gupta NP, Kucheria K, Gupta SK, Screening for Y-chromosome microdeletions in infertile Indian males: Utility of simplified multiplex PCRIndian J Med Res 2008 127(2):124-32. [Google Scholar]
[68]. Dada R, Gupta NP, Kucheria K, Cytogenetic and molecular analysis of male infertility: Y chromosome deletion during nonobstructive azoospermia and severe oligozoospermiaCell Biochem Biophys 2006 44:171-77.10.1385/CBB:44:1:171 [Google Scholar] [CrossRef]
[69]. Sachdeva K, Saxena R, Majumdar A, Chadda S, Verma IC, Use of ethnicity-specific sequence tag site markers for Y chromosome microdeletion studiesGenet Test Mol Biomarkers 2011 15:451-59.10.1089/gtmb.2010.015921375402 [Google Scholar] [CrossRef] [PubMed]
[70]. Esteves SC, Miyoka R, Agarwal A, An update on the clinical assesment of the infertile maleClinics 2011 66(4):691-700.10.1590/S1807-5932201100040002621655766 [Google Scholar] [CrossRef] [PubMed]
[71]. Barchi M, Roig I, Di Giacomo M, De Rooij DG, Keeney S, Jasin M, ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomesPLoS Genet 2008 4:e100007610.1371/journal.pgen.100007618497861 [Google Scholar] [CrossRef] [PubMed]
[72]. Vogt P, Keil R, Köhler M, Lengauer C, Lewe D, Lewe G, Selection of DNA sequences from interval 6 of the human Y chromosome with homology to a Y chromosomal fertility gene sequence of Drosophila hydeiHuman Genetics 1991 86(4):341-49.10.1007/BF002018301999335 [Google Scholar] [CrossRef] [PubMed]
[73]. Kleiman SE, Almog R, Yogev L, Hauser R, Lehavi O, Paz G, Screening for partial AZFa microdeletions in the Y chromosome of infertile men: Is it of clinical relevance?Fertil Steril 2012 98:43-47.10.1016/j.fertnstert.2012.03.03422537385 [Google Scholar] [CrossRef] [PubMed]
[74]. Wei W, Fitzgerald T, Ayub Q, Massaia A, Smith BB, Dominiczak AA, Copy number variation in the human Y chromosome in the UK populationHum Genet 2015 134:789-800.10.1007/s00439-015-1562-525957587 [Google Scholar] [CrossRef] [PubMed]
[75]. Nailwal M, Chauhan JB, Azoospermia factor a (AZFa) sub-region of human Y- chromosome: A reviewMeta Gene 2017 13:124-28.10.1016/j.mgene.2017.06.001 [Google Scholar] [CrossRef]
[76]. Hotaling JM, Smith JF, Rosen M, Muller CH, Walsh TJ, The relationship between isolated teratozoospermia and clinical pregnancy after in vitro fertilization with or without intracytoplasmic sperm injection: A systematic review and meta-analysisFertility and Sterility 2011 95(3):1141-45.10.1016/j.fertnstert.2010.09.02921030014 [Google Scholar] [CrossRef] [PubMed]
[77]. Laaser I, Theis FJ, de Angelis MH, Kolb HJ, Adamski J, Huge splicing frequency in human Y chromosomal UTY geneOmics: A Journal of Integrative Biology 2011 15(3):141-54.10.1089/omi.2010.010721329462 [Google Scholar] [CrossRef] [PubMed]
[78]. Simoni M, Tüttelmann F, Gromoll J, Nieschlag E, Clinical consequences of microdeletions of the Y chromosome: The extended Münster experienceReproductive Biomedicine Online 2008 16(2):289-303.10.1016/S1472-6483(10)60588-3 [Google Scholar] [CrossRef]
[79]. Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN, Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regionsHum Reprod 2003 18(8):1660-65.10.1093/humrep/deg34812871878 [Google Scholar] [CrossRef] [PubMed]
[80]. Abid S, Maitra A, Meherji P, Patel Z, Kadam S, Shah J, Clinical and laboratory evaluation of idiopathic male infertility in a secondary referral center in IndiaJ Clin Lab Anal 2008 22:29-38.10.1002/jcla.2021618200580 [Google Scholar] [CrossRef] [PubMed]
[81]. Simoni M, Gromoll J, Dworniczak B, Rolf C, Abshagen K, Kamischke A, Screening for deletions of the Y chromosome involving the DAZ (Deleted in AZoospermia) gene in azoospermia and severe oligozoospermiaFertility and Sterility 1997 67(3):542-47.10.1016/S0015-0282(97)80083-0 [Google Scholar] [CrossRef]
[82]. Krausz C, Degl’Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, Natural transmission of USP9Y gene mutations: A new perspective on the role of AZFa genes in male fertilityHuman Molecular Genetics 2006 15(18):2673-81.10.1093/hmg/ddl19816893908 [Google Scholar] [CrossRef] [PubMed]
[83]. Fu L, Xiong DK, Ding XP, Li C, Zhang LY, Ding M, Genetic screening for chromosomal abnormalities and Y chromosome microdeletions in Chinese infertile menJournal of Assisted Reproduction and Genetics 2012 29(6):521-27.10.1007/s10815-012-9741-y22415247 [Google Scholar] [CrossRef] [PubMed]
[84]. Krausz C, Hoefsloot L, Simoni M, Tüttelmann F, EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: State-of-the-art 2013Andrology 2014 2(1):05-19.10.1111/j.2047-2927.2013.00173.x24357628 [Google Scholar] [CrossRef] [PubMed]
[85]. Bunyan DJ, Callaway JL, Laddach N, Detection of partial deletions of Y- chromosome AZFc in infertile men using the multiplex ligation-dependent probe amplification assayJ Reprod Infertil 2012 13(3):174-78. [Google Scholar]
[86]. Osborne EC, Lynch M, Melachlan R, Trouns Ao, Cram DS, Microarray deletion of Y chromosome deletions associated with male infertilityRBM Online 2007 15:623-80.10.1016/S1472-6483(10)60534-2 [Google Scholar] [CrossRef]
[87]. van Golde RJ, Wetzels AM, de Graaf R, Tuerlings JH, Braat DD, Kremer JA, Decreased fertilization rate and embryo quality after ICSI in oligozoospermic men with microdeletions in the azoospermia factor c region of the Y chromosomeHuman Reproduction 2001 16(2):289-92.10.1093/humrep/16.2.28911157822 [Google Scholar] [CrossRef] [PubMed]
[88]. Gonçalves C, Cunha M, Rocha E, Fernandes S, Silva J, Ferraz L, Y-chromosome microdeletions in nonobstructive azoospermia and severe oligozoospermiaAsian Journal of Andrology 2017 19(3):33810.4103/1008-682X.17282726908064 [Google Scholar] [CrossRef] [PubMed]
[89]. Aitken RJ, Krausz C, Oxidative stress, DNA damage and the Y chromosomeReproduction-Cambridge- 2001 122(4):497-506.10.1530/reprod/122.4.49711570956 [Google Scholar] [CrossRef] [PubMed]
[90]. Agarwal A, Said TM, Role of sperm chromatin abnormalities and DNA damage in male infertilityHum Reprod Update 2003 9:331-45.10.1093/humupd/dmg02712926527 [Google Scholar] [CrossRef] [PubMed]
[91]. Plastira K, Msaouel P, Angelopoulou R, Zanioti K, Plastiras A, Pothos A, The effects of age on DNA fragmentation, chromatin packaging and conventional semen parameters in spermatozoa of oligoasthenoteratozoospermic patientsJ Assist Reprod Genet 2007 24:437-43.10.1007/s10815-007-9162-517768675 [Google Scholar] [CrossRef] [PubMed]
[92]. Seli E, Gardner DK, Schoolcraft WB, Moffatt O, Sakkas D, Extent of nuclear DNA damage in ejaculated spermatozoa impacts on blastocyst development after in vitro fertilizationFertil Steril 2004 82:378-83.10.1016/j.fertnstert.2003.12.03915302287 [Google Scholar] [CrossRef] [PubMed]
[93]. Larson-Cook KL, Brannian JD, Hansen KA, Kasperson KM, Aamold ET, Relationship between the outcomes of assisted reproductive techniques and sperm DNA fragmentation as measured by the sperm chromatin structure assayFertility and Sterility 2003 80(4):895-902.10.1016/S0015-0282(03)01116-6 [Google Scholar] [CrossRef]
[94]. Athalye AS, Madon PF, Naik NJ, Naik DJ, Gavas SS, Dhumal SB, A study of Y chromosome microdeletions in infertile Indian malesInt J Hum Genet 2004 4:179-85.10.1080/09723757.2004.11885889 [Google Scholar] [CrossRef]
[95]. Waseem AS, Singh V, Makker GC, Trivedi S, Mishra G, Singh K, AZF deletions in Indian populations: Original study and meta-analysesJournal of Assisted Reproduction and Genetics 2020 37(2):459-69.10.1007/s10815-019-01661-031919744 [Google Scholar] [CrossRef] [PubMed]