Pyogenic infections characterised by inflammation with exudate formation. Pyogenic infection is a serious threat leading to sepsis. The most important cause of pyogenic infections is Staphylococcus aureus followed by gram negative organisms [1]. The source of infection could be either endogenous or exogenous. It usually begins with the break in the epithelial barrier of the skin which gradually allows the organism in the skin to proliferate slowly and cause infection. The defense mechanism of our body fights by assembling the immune cells to the site of inflammation. Eventually the accumulation of these cells leads to formation of pus [2,3]. Studies in India show a different situation where the gram negative pathogens predominate the gram positive pathogens in causing pyogenic infections [4,5]. The unscrupulous administration of antibiotics has caused the emergence of MDR organisms thus making their treatment difficult. In this scenario where the infections are a major cause of morbidity and mortality, it is always a mandate to know the causative organism and the susceptibility profile to commence the right treatment for the patient at the earliest [4,5].
Various studies about the bacterial profile involved in causing pyogenic infections are available around the globe [1,2,4,5]. But since the susceptibility of the organisms to the antibiotics keeps changing a continuous surveillance of the susceptibility pattern is warranted to identify the present situation. Therefore, this study was undertaken to identify the aerobic organisms involved in causing various pyogenic infections with their current susceptibility pattern.
Materials and Methods
A retrospective analysis was carried out on 750 culture positive pus and wound swabs which was received in the Department of Microbiology from various departments of the hospital (Surgery, Orthopaedics, ENT, Ophthal, Obstetrics and Gynaecology, Neurosurgery, Cardiothoracic Surgery and Surgical Gasteroenterology) between June 2018-June 2019. The data regarding the organism profile and their susceptibility pattern was collected from the records.
Microbiological Work-up
All pus and wound swabs that were received in the microbiology department for culture and sensitivity were processed by standard methods [6]. All samples were cultured on the 5% sheep blood agar and Macconkey agar and incubated at 37°C for 24-48 hours in aerobic environment. The colonies were subjected to gram staining, motility testing and biochemical tests used for identification like catalase, coagulase, oxidase, indole production, citrate utilisation, urease production, triple sugar iron, mannitol motility medium and fermentation of sugars like glucose, sucrose, lactose and mannitol. Simultaneously, the individual colonies were also used to perform the Antimicrobial Susceptibility Testing (AST) using Kirby-Bauer method of disc diffusion on Muller-Hinton medium according to CLSI guidelines (2018) [7]. For the detection of MRSA, disc diffusion method was employed with cefoxitin (30μg) disc according to CLSI guidelines. All the media and antibiotic used were obtained from Hi media.
For gram positive cocci-penicillin (10units), erythromycin (15 μg), clindamycin (2 μg), ciprofloxacin (5 μg), gentamicin (10 μg), linezolid (30 μg), teicoplanin (30 μg), high level gentamicin (120 μg) in case of Enterococcus were used.
For gram negative bacilli-gentamicin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), ceftriaxone (30 μg), cephotaxime (30 μg), ceftazidime (30 μg), imipenem (10 μg), piperacillin tazobactam (100 μg), cefoperazone-sulbactam (100 ug), meropenem (10 μg), tigecycline (15 ug) were used. For colistin, microbroth dilution method was used [7].
ESBL was detected by combined disk test. This was performed by phenotypic confirmatory test as per the recommendations of CLSI [7]. The ceftazidime (30 μg) discs alone and in combination with clavulanic acid (ceftazidime +clavulanic acid, 30/10 μg discs) were used. An increase of ≥5 mm in zone of inhibition of the combination discs in comparison to the ceftazidime disc alone was considered to be ESBL producer.
MRSA was detected by cefoxitin disc diffusion test. Lawn culture was done onto Mueller-Hinton agar plate. A 30 μg cefoxitin disc was placed and incubated at 37°C for 24 hours. The zone of inhibition of S. aureus ≤21 mm was considered as methicillin resistant [7].
Statistical Analysis
The results thus obtained were analysed using Statistical Package for the Social Sciences (SPSS) software (version 21) and were represented in the form of percentages and frequencies.
Results
Among the total 942 samples obtained from various department of Karpagam Faculty of Medical Sciences and Research Hospital, Coimbatore, Tamil Nadu, India, for aerobic culture and sensitivity, 750 (79.6%) samples were positive by culture for either gram positive or gram negative organisms. About 192 (20.4%) samples showed no growth. The samples were obtained from variety of cases like cellulitis (n=108), diabetic ulcers (n=302), postoperative wound infections (n=209), skin and soft tissue infections (n=243), secondary infection in eczema patients (n=33), folliculitis (n=25), empyema (n=22). There were 30 people from less than 20 years of age group, 465 people between 21 and 40 years of age, 301 people between 41 and 60 years, 112 members between 61 and 80 and 34 members over 80 years of age. Frequency of bacterial isolation was maximum among age group 21 to 40 years followed by 41 to 60 years.
Among the 750 positive samples analysed, males were 523 while females were 227. Males outnumbered females (M:F-2.3:1) and the median age was 49 years [Table/Fig-1].
Gender wise distribution of patients and samples.
| Gender | Total number of samples collected (n=942) | Total number of culture positives (n=750) |
|---|
| Male | 652 | 523 |
| Female | 290 | 227 |
| Total | 942 | 750 |
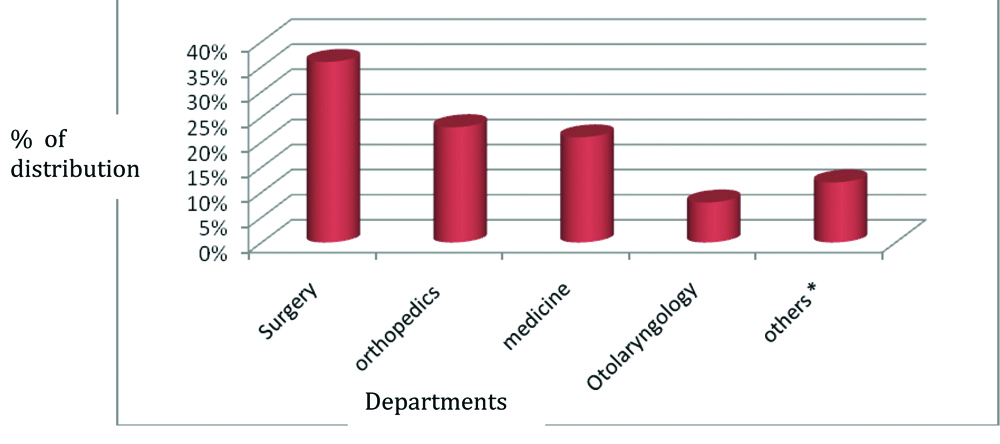
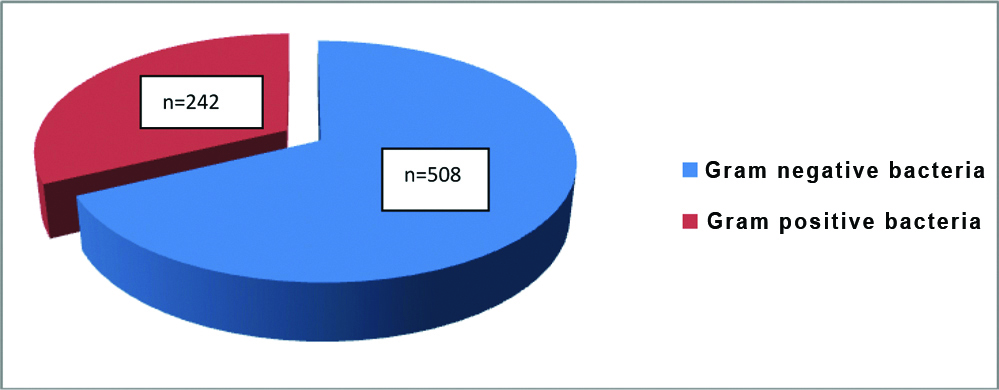
Among the various departments that submitted the samples, major contribution was from department of surgery (36%) followed by orthopaedics (23%), medicine (21%), otolaryngology (8%) and others (12%) [Table/Fig-2]. The number of gram positive bacteria isolated was 242/750 (32.3%) and gram negative organisms were 508/750 (67.7%) [Table/Fig-3].
Department wise distribution of samples.
*paediatrics, dermatology, ophthalmology, obstetrics and gynaecology
Percentage of gram positive and gram negative bacteria isolated from pus/wound swabs.
In present study the percentage of monomicrobial infection was 637/750 (85%) and polymicrobial infections were 113/750 (15%). The percentage of MDR was high among gram negatives than gram positives.
The most common causative agent for pyogenic infection according to present study was Pseudomonas aeruginosa (17.7%) (n=133/750). The predominant gram negative bacteria were Pseudomonas aeruginosa while others were Escherichia coli, Proteus mirabilis, Enterobacter spp, Klebsiella pneumonia [Table/Fig-4]. The predominant gram positive bacteria isolated were MRSA and MSSA [Table/Fig-5]. There was no fungal isolates identified in the study.
Number of gram negative organisms isolated (n=508/750).
| S. No. | Organism | Total | Percentage |
|---|
| 1 | Pseudomonas aeruginosa | 133 | 26.1% |
| 2 | Escherichia coli | 128 | 25.1% |
| 3 | Proteus mirabilis | 72 | 14.1% |
| 4 | Enterobacter spp | 48 | 9.4% |
| 5 | Klebsiella pneumoniae | 42 | 8.2% |
| 6 | Morganella morgagni | 23 | 4.5% |
| 7 | Non-fermenting Gram negative bacilli | 20 | 3.9% |
| 8 | Proteus vulgaris | 14 | 2.7% |
| 9 | Acinetobacter baumanii | 9 | 1.7% |
| 10 | Klebsiella oxytoca | 7 | 1.3% |
| 11 | Providencia spp | 5 | 0.9% |
| 12 | Citrobacter koseri | 2 | 0.3% |
| 13 | Citrobacter diversus | 2 | 0.3% |
| 15 | Citrobacter fruendi | 1 | 0.1% |
| 14 | Citrobacter spp (other) | 1 | 0.1% |
| 16 | Serratia marcesens | 1 | 0.1% |
Number of gram positive organisms isolated (n=242/750).
| S. No. | Organism | Total | Percentage |
|---|
| 1 | MRSA | 100 | 41.3% |
| 2 | MSSA | 80 | 33% |
| 3 | MRCONS | 19 | 7.8% |
| 4 | Enterococcus spp | 28 | 11.5% |
| 5 | MSCONS | 6 | 2.4% |
| 6 | Streptococcus viridans | 3 | 1.2% |
| 7 | Streptococcus spp | 3 | 1.2% |
| 8 | Beta Haemolytic Strepotococcus spp | 3 | 1.2% |
MRSA: Methicillin-resistant staphylococcus aureus; MSSA: Methicillin-sensitive staphylococcus aureus; MRCONS: Methicillin-resistant coagulase negatine staphylococci; MSCONS: Methicillin-sensitive coagulase negatine staphylococci
Among the gram negatives 117/508 (23%) were ESBL producers and majority of them were Escherichia coli (56/117) [Table/Fig-6]. MDR was seen in 6/508 in gram negatives among which majority of them were Klebsiella pneumoniae and NFGNB [Table/Fig-7]. Antibiotic resistance pattern of Staphylococcus aureus and Pseudomonas aeruginosa is shown in [Table/Fig-8,9].
List of ESBL producers isolated from wound swabs and pus.
| S. No. | ESBL Producers (n=117/508) | Percentage |
|---|
| 1 | Escherichai coli (n=56) | 47.8% |
| 2 | Klebsiella pneumonia (n=14) | 11.9% |
| 3 | Klebsiella oxytoca (n=8) | 6.8% |
| 4 | Proteus mirabilis (n=14) | 11.9% |
| 5 | Proteus vulgaris (n=1) | 0.8% |
| 6 | Acinetobacter baumani (n=2) | 1.7% |
| 7 | NFGNB (n=9) | 7.6% |
| 8 | Providentia spp (n=1) | 0.8% |
| 9 | Pseudomonas aeruginosa (n=12) | 10.2% |
NFGNB: Non-fermenting gram negative bacilli
List of gram negative MDR pathogens isolated.
| S. No. | MDR Pathogens (n=6) | Percentage |
|---|
| 1 | Klebsiella pneumonia (n=2) | 33.3% |
| 2 | NFGNB (n=2) | 33.3% |
| 3 | Pseudomonas aeruginosa (n=1) | 16.6% |
| 4 | Escherichia coli (n=1) | 16.6% |
NFGNB: Non-fermenting gram negative bacilli
Antibiotic resistance pattern in Staphylococcus aureus.
| Resistogram | Antibiotics | Resistance strains (N=180) | Percentage |
|---|
| Staphylococcus aureus | Penicillin | 164 | 91.1% |
| Erythromycin | 80 | 44% |
| Ciprofloxacin | 62 | 34.4% |
| Gentamycin | 70 | 38.8% |
| Cefoxitin | 100 | 55.5% |
| Linezolid | 1 | 0.5% |
| Clindamycin | 53 | 29.4% |
Antibiotic resistance pattern in Pseudomonas aeruginosa.
| Resistogram | Antibiotics | Resistance strains (N=133) | Percentage |
|---|
| Pseudomonas aeruginosa | Amikacin | 49 | 36.8 |
| Gentamicin | 58 | 43.6 |
| Ceftazidime | 41 | 30.8 |
| Ceftrioxone | 33 | 24.8 |
| Cefotaxime | 31 | 23.3 |
| Cefoperazonesulbactam | 41 | 30.8 |
| Piperacillin tazobactam | 11 | 8.2 |
| Ciprofloxacin | 24 | 18 |
| Imipenem | 19 | 14.2 |
| Meropenem | 15 | 11.2 |
| Colistin | 0 | 0 |
| Tigecycline | 9 | 6.7 |
Discussion
This study was conducted in Department of Microbiology in a tertiary care hospital. Pus samples received from different departments of our hospital were 942, of which 750 (79.6%) gave a positive aerobic bacterial growth. The present study was conducted in a tertiary hospital which showed a predominance of gram negative organisms in the skin and soft tissue infections.
Monomicrobial/Polymicrobial
The cause of the infection may be monomicrobial or polymicrobial [8]. In present study majority of the infections were monomicrobial (85%) accounting for 637 out of 750 samples which were culture positive. The polymicrobial infection rate was 15%. Combined infections which were polymicrobial were also noted by Ananthi B et al., in their study [9]. The severity of the infection decides the length of stay of the person in the hospital thus increasing the overall cost and burden of the patient.
Gram Negative Predominance
In present study gram negative bacilli was predominant accounting for 67.7% while the gram positive bacteria were 32.3% which again correlates with the changing trend in India with gram negative organism being predominant than gram positive organisms as discussed by study from Southern India, Krishnamurthy S et al., [5]. The results obtained were similar to observations made by Mahat P et al., and Yakha JK et al., from Northern India [10,11], where the percentage of gram negative organisms were 71.82% and gram positives were 28.18% [10] and 70.6% and 29.4% [11] respectively.
The predominant gram negative bacteria isolated in present study was Pseudomonas aeruginosa (26.1%) followed by Escherichia coli (25.1%). This observation was against the finding of Krishnamurthy S et al., where Klebsiella pneumonia was the commonest organism isolated from pus samples [5].
Over the past few years there is an increase in gram negative organisms causing pyogenic infections than the gram positive organisms. This trend has been documented in many studies from 2013 onwards [Table/Fig-10] [5,10-13]. The similar findings have re-emphasised the predominance of gram negative pyogenic infections.
Evidence of increasing incidence of gram negative pyogenic infections.
| Studies | Year | Place | Sample size | Prevalence |
|---|
| Mahat P et al., [10] | 2017 | Kathmandu, Nepal | 503 | Pseudomonas spp. (34.55%) followed by Staphylococcus aureus (21.36%) [10] |
| Krishnamurthy S et al., [5] | 2016 | Karimnagar, Telangana, India | 383 | Klebsiella pneumoniae (34.46%) followed by Staphylococcus aureus (18.53%) [5] |
| Yakha JK et al., [11] | 2014 | Lalitpur, Nepal | 870 | 70.6% were Gram-negative and 29.4% were gram-positive [11] |
| Sharma V et al., [12] | 2015 | Ajmer, Rajasthan | 100 | Klebsiella spp (28%) followed by Pseudomonas aeruginosa 20 (20%) [12] |
| Panta K et al., [13] | 2013 | Kathmandu, Nepal | 1110 | E. coli (113/181) was the major followed by Salmonella Typhi (17/181) [13] |
| Gunasekaran J et al., (present study) | 2020 | Coimbatore, India | 942 | 67.7% were gram-negative and 32.3% were gram-positive. Pseudomonas aeruginosa was the predominant gram negative organism |
β-lactamases, which are responsible for resistance of β-lactam group of antibiotics, hydrolyse the amide bond of the four-membered characteristic β-lactam ring, thus rendering the antimicrobial ineffective [14]. In this study, though Pseudomonas spp. was most commonly isolated, the percentage of ESBL producing strains among them were only 10.2% and MDR was (16.6%). Analysing the resistogram of Pseudomonas aeruginosa isolates revealed that highest resistance was shown to gentamicin (43.6%) followed by amikacin (36.8%). Resistance to beta lactam and beta lactamase inhibitor drugs like cefoperazone sulbactam and piperacillin tazobactam were 30.8% and 8.2% in present study. Resistance due to carbapenemase was found to be slightly more for imipenem than meropenem in our isolates. The total number of ESBL producers in the study was observed to be 23% whereas the study done by Wadekar MD et al., observed the trend to be around 61.2% [14].
Escherichia coli were seen in 128 samples accounting for 25.1% next to Pseudomonas aeruginosa. The resistance pattern of Escherichia coli was alarming as almost 47.8% of them were ESBL producers and 16.6% were MDR. Though isolated a little less commonly than Pseudomonas aeruginosa, Escherichia coli infections were severe thus increasing the burden to the patient due to increased hospital stay. Another study showed that there is an increase in isolation of ESBL Esherichia coli from pus samples over a period of time from 2005 to 2010 from 41.1% to 55.5% [15].
Other Enterobacteriaceae members like Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis and vulgaris, Providencia spp were isolated in present study at the rate of 8.2%, 1.3%, 14.1%, 2.7%, and 0.9%, respectively. Other Enterobacteriaceae members that were rarely isolated were Morganella spp, Citrobacter spp, and Serratia spp. Klebsiella spp were the second common ESBL producers with K.pneumoniae contributing to 11.9% and K.o
xytoca accounting to 6.8%. Non-fermenting gram negative bacteria were isolated in the rate of 3.9%. Acinetobacter baumani was isolated at the rate of 1.7%. Similar organisms were isolated in the study done by Wadekar MD et al., [14]. The rate of ESBL production by NFGNB was 7.6% and MDR was 33.3%. These gram negative organisms which are commonly found in hospital environment, tend to be resistant to common antiseptics and are also MDR.
In a study done by Magiorakos AP et al., they found that the prevalence of ESBL producing organisms was found 18% amongst which E. coli was 53.7%, K. pneumoniae 14.8%, P. mirabilis 12.9% and others 7.4% [16]. In present study, that ESBL producing E.coli was 47.8% followed by Klebsiella spp (18.9%), Proteus spp (12.8%), Non-fermenters like Pseudomonas spp, Acinetobacter spp and others contributed to 20.5%.
MDR is the resistance to more than three classes of antibiotics of the five classes of antibiotics like beta lactam/beta lactamase inhibitor combinations, 3rd generation cephalosporins, carbapenems, fluoroquinolones and aminoglycosides [17-19]. In a study in 2016, it was observed that MDR E. coli was found in 31.6%, followed by K. pneumoniae 30% [16]. In present study, Klebsiella pneumoniae and NFGNB as predominant MDR organisms contributing to 33.3%. colistin and tigecycline was used for the treatment of patients suffering from MDR organisms. There was no fungus isolated in present study.
Among the 242 gram positive bacterial isolates isolated majority of them Staphylococcusaureus was the predominant. It is the most common isolate in most of the studies [20-22]. In a study done in Bangalore; S. aureus was in 40.5% of wound infections [1]. Bowler PG et al., in their study showed S.aureus was the most common cause of cutaneous abscess and mostly associated with acute soft tissue infections [23]. It is also associated with diabetic wound ulcers and delayed wound infection [16,23]. In present study Staphylococcusaureus was seen in 180 of 242 gram positive gram positive organisms isolated (74.3%). Out of these, predominant was MRSA (41.3%) and MSSA were 33%. MRSA strains are organisms that are resistant to a large group of antibiotics having the beta-lactams ring, including penicillins and cephalosporins. Methicillin resistance is rendered to the bacteria by the acquisition of a mecA gene. This produces an alternative Penicillin Binding Protein 2a (PBP2a), which has lower affinity for β-lactam antibiotics [14,24].
Other gram positive organisms isolated were Enterococcus spp, Beta haemolytic Streptococcus spp and CONS as seen by Verma P, where the common organisms isolated were Staphylococcus and Streptococcus spp [25]. The resistogram of Staphylococcus aureus showed that penicillin was resistant in 91% of isolates followed by macrolide erythromycin which was 44%, Gentamicin was 38.8% and clindamycin was 29.4%. Cefoxitin was used as a surrogate marker for the detection of MRSA where almost 25% of isolates were resistant to it.
In the present study, piperacillin tazobactam (90%), meropenem (83%) and imipenem (87%) were the antibiotics to which most of the gram negative organisms were susceptible which was similar to study done by Wadekar MD et al., and Rameshkannan S et al. and Cardoso T et al., which showed maximum susceptibility to these antibiotics [14,26,27]. Similarly most of gram positive isolates were sensitive to linezolid (98%) which is same as the results of studies conducted by Verma P and Shittu AO et al., [25,28].
It can be concluded that monomicrobial infections are more common with a significant predominance of gram negative infection (Pseudomonas aeruginosa followed by Escherichia coli), which is a changing trend in India. The MDR organisms like Pseudomonas aeruginosa and MRSA contributed to majority of cases in present study observation. Piperacillin tazobactam and carbapenems were effective drugs for most of the gram negatives and linezolid was effective for the gram positive organisms.
Limitation(s)
As present study was a retrospective study, certain factors like source of infection, the duration of hospital stay, and clinical outcome were not considered in the evaluation.
Conclusion(s)
Present study was done in a tertiary care centre major group of patients get admitted after getting treated from outside hospitals. Most of them were treated with higher class of antibiotics in other hospitals which may have lead to the growth of MDR pathogens. Diligent control over the usage of antibiotics and infection control measures in every level would go a long way in the control of infection with resistant pathogens. Since the sensitivity pattern is different in different regions of the country there must be a proper institutional antibiogram and formulation of antibiotic policy would help in empiric antibiotic therapy with a reduction infection rates.
MRSA: Methicillin-resistant staphylococcus aureus; MSSA: Methicillin-sensitive staphylococcus aureus; MRCONS: Methicillin-resistant coagulase negatine staphylococci; MSCONS: Methicillin-sensitive coagulase negatine staphylococci
NFGNB: Non-fermenting gram negative bacilli
NFGNB: Non-fermenting gram negative bacilli