Carbapenem Resistant Enterobacteriaceae (CRE) is a group of multidrug-resistant bacteria which is increasingly reported worldwide [1]. Various infections caused by CRE e.g., bloodstream infection, respiratory infections, urinary tract infections, etc., are difficult to treat due to extensive drug resistance to most of the antimicrobial agents used routinely. Rapid and accurate detection of CRE is the need of time. Due to the presence of more than one mechanism of resistance, there is considerable heterogeneity present in every method for its detection. CREs differed from each other by either producing carbapenemase enzyme and called Carbapenemase Producing Carbapenem Resistant Enterobacteriaceae (CP-CRE) or Non-Carbapenemase Producing Carbapenem Resistant Enterobacteriaceae (nonCP-CRE) [2]. Clinical and Laboratory Standards Institute (CLSI) defines an isolate to be CRE based on either by the demonstration of resistance to any of the carbapenem (imipenem, meropenem, doripenem, or ertapenem) by disc diffusion/determination of Minimum Inhibitory Concentration (MIC) breakpoints and/or proven to have carbapenemase enzyme by phenotypic tests such as CNP test [3-5]. A disc diffusion test is observed to be a reliable method for any kind of carbapenem resistance and is used as a screening test for CRE detection [6]. Ertapenem among the carbapenem discs has been observed as a marker for CRE detection, primarily caused by the mechanism other than carbapenemase production, such as production of AmpC beta-lactamases/Extended-spectrum of Beta-Lactamase (ESBL) with loss of porin channels and over expression of efflux pumps, etc., [7-11]. There are very few literature showing the role of ertapenem in detecting CRE other than carbapenemases and the sensitivity and specificity of the ertapenem disc diffusion test compared to gold standard tests.
The current study aimed to evaluate the role of ertapenem disc as a marker for detecting CRE with respect to other discs individually and in combination, where E tests and the gold standard tests i.e., CNP test, and PCR were also compared. Authors also analysed its usage for the differentiation between CP-CRE and nonCP-CRE.
Materials and Methods
A prospective study was carried out from January to December 2017 over a period of one year to find the concordance of susceptibility by ertapenem disc with other carbapenem discs (i.e., imipenem, meropenem, and doripenem) by disc diffusion test, E strip method and other gold-standard tests such as CNP and PCR and to determine the role of non-susceptibility of ertapenem for the detection and differentiation between CP-CRE and nonCP-CRE. The study was approved by the Institutional Ethical Committee (Ref no- IECPG-157/27.01.2016). Informed consent was taken from all patients who participated in the study.
A total of 76 Enterobacteriaceae non-repetitive isolates from the rectal swab of admitted patients following the Centre for Disease Control and Prevention (CDC) protocol were included in the study [12]. All the isolates were confirmed for its identification by Matrix-Assisted Laser Desorption Ionisation-Time Of Flight Mass Spectrometry (MALDI-TOF MS, VITEK-MS system, BioMérieux, Marcy-l’Étoile, France). All the isolates were first screened for carbapenem resistance using the disc diffusion method by Kirby Bauer method {Imipenem (10 μg), meropenem (10 μg), doripenem (10 μg), and ertapenem (10 μg), HiMedia, Mumbai}. Isolates other than Enterobacteriaceae were excluded. The antibiotic discs were kept at 2-8°C temperature, and quality control was done twice in a week. E tests with predefined antibiotic gradients (ranges from 0.002-32 μg/mL) were used for the corresponding carbapenems to determine MIC breakpoints. The interpretation of the result of disc diffusion test and MIC breakpoints was done as per the CLSI 2017 guideline [3]. All the isolates were further tested for the CNP test and carbapenemase genes (blaNDM-1, blaOXA-48, blaKPC, blaVIM, blaIMP) (New Delhi Metallo-beta-lactamase (NDM), Oxacillin-hydrolyzing beta-lactamase (OXA) Klebsiella pneumoniae carbapenemase (KPC), Verona imipenemase (VIM) and Imipenemase (IMP)) by conventional PCR using published primers [3,8]. The CRE detection by either CNP or PCR was considered as the gold standard.
Molecular Analysis
All the isolates were subjected to PCR analysis. Deoxyribonucleic Acid (DNA) extraction for all the CRE isolates was completed and stored at -20°C for further analysis. The PCR for blaNDM-1, blaKPC,blaOXA-48, blaIMP, and blaVIM gene had been standardised using primers from the published literature [Table/Fig 1] [8,13]. The primer sequences are mentioned below.
Details of primers sequences for gene blaNDM-1, blaKPC, blaOXA-48, blaIMP, and blaVIM.
| Gene | Nucleotides sequence (5’-3’) | Size of the product (bp) |
|---|
| NDM-1-F | GGTGCATGCCCGGTGAAATC | 660 |
| NDM-1-R | ATGCTGGCCTTGTTTAACG |
| KPC-F | ATGTCACTGTATCGCCGTC | 382 |
| KPC-R | AATCCCTCCGAGCGCGAGT |
| OXA-48-F | GCGTGGTTAAGGATGAACAC | 438 |
| OXA-48-R | CATCAAGTTCAACCCAACCG |
| IMP-F | GGCAGTCGCCCTAAAACAAA | 737 |
| IMP-R | TAGTTACTTGGCTGTGATGG |
| VIM-F | AAAGTTATGCCGCACTCACC | 865 |
| VIM-R | TGCAACTTCATGTTATGCCG |
(New Delhi Metallo-beta-lactamase (NDM), Oxacillin-hydrolyzing beta-lactamase (OXA) Klebsiella pneumoniae carbapenemase (KPC), Verona imipenemase (VIM) and Imipenemase (IMP), F: Forward, R: Reverse
The PCR amplification was performed in a 25 μL reaction volume. A 3 μL genomic DNA was added to the PCR reaction mixture containing 10 μM primer concentration of each primer and 1.25 UTaq- DNA polymerase. The PCR cycling protocol involved an initial 10 minutes denaturation step at 95°C followed by 35 cycles of 45 seconds of denaturation at 94°C, 45 seconds of primer annealing at respective temperature, and 50 seconds of primer extension at 72°C. Following the single subsequent elongation step at 72°C for a 7 minute primer extension, the products were held at 4°C. Then, gel electrophoresis was performed with 1% agarose and ethidium bromide, and bands were observed in the amplified product on UV transilluminator.
Commercial CNP (RAPIDECR CARBA NP, BioMerieux) was used, and the result was interpreted according to the manufacturer’s instructions. After two hours of incubation, change of colour from red to yellow was considered as test positive, whereas no change of colour or change from red to orange was considered as test negative.
Analysis
The analysis was made based on the result of the disc diffusion test. To evaluate the potency of ertapenem disc in comparison to other carbapenem discs, the isolates were tested for disc diffusion test and assessed in the following manner: 1) Isolates resistant to any one of the carbapenem disc apart from ertapenem; 2) Isolates resistant to any two of the carbapenem disc apart from ertapenem; 3) Isolates resistant to all the three carbapenem discs apart from ertapenem; 4) Isolates resistant to ertapenem disc only. Simultaneously, corresponding antibiotic E strip tests were tested to determine the breakpoints for all the isolates. Concordance of the antibiotic susceptibility result by disc diffusion and E strip method for all the carbapenem drugs were compared. Finally, the susceptibility result of ertapenem disc was compared with the gold standard PCR and CNP test. For the detection of CP-CRE, the following criteria were followed-up [3,14].
True positive for CP-CRE: Isolates found positive for carbapenemase production by either CNP test and/or PCR and resistant by disc diffusion test.
False-negative for CP-CRE: Isolates produce carbapenemase by CNP test and/or PCR but sensitive by disc diffusion test.
False-positive for CP-CRE: Isolates negative for carbapenemase production by either CNP test and or PCR and resistant by disc diffusion test.
True negative for CP-CRE: Isolates negative for carbapenemase production by either CNP test and/or PCR and by disc diffusion test.
Statistical Analysis
Data were analysed using Statistical Package for the Social Sciences (SPSS) software v.20.0 (SPSS Inc., Chicago, IL) by χ2 test. Significance was set at p<0.05 using two-sided comparisons.
Results
A total of 76 non-repetitive isolates were included. On comparing the demographic data, males were the predominant group {75%, (57/76)} followed by females {25%, (19/76)} with 60.5% adult population. Mean age distribution among adult and paediatrics patients were 27 years (18-46 years) and 9 years (10 months-17 years), respectively. In this pilot study, a total of 76 phenotypically confirmed Enterobacteriaceae isolates form rectal swab were tested for the disc diffusion, E test, CNP, and PCR. By CNP 58 were positive, 17 were negative, and one was indeterminate (positive by PCR). Total 52 isolates were positive and 24 were negative by PCR. PCR for blaNDM-1 was positive in 38 isolates [Table/Fig-2a], blaOXA-48 was positive in 24 isolates [Table/Fig-2b], both blaNDM-1 and blaOXA-48 were positive in 10 isolates and blaIMP was positive in 1 isolate [Table/Fig-2c]. None of the isolates were positive for blaKPC and blaVIM. Five isolates were negative by both the tests. So, using the above criteria, 68 out of 76 were classified as CP-CRE, and five were nonCP-CRE. Three isolates were found sensitive to all the carbapenems by disc diffusion and E test, CNP, PCR, and were considered as non-CREs. Among all the carbapenem discs, ertapenem alone detected the maximum number of CRE isolates (84.2%) followed by others when compared singly or with the combination [Table/Fig-3,4]. This proves the higher sensitivity of ertapenem disc compared to other carbapenem discs alone and in combination for the detection of CRE. We also tried to compare the concordance or discordance of the result between the disc diffusion test methods with that of its corresponding E tests. Among the four carbapenems, maximum concordance of susceptibility result was observed between the ertapenem disc and the ertapenem E test [Table/Fig-5]. In 55 isolates, both the DD and E test were resistant using ertapenem in comparison to 18, 23 and 18 isolates by imipenem, meropenem and doripenem, respectively. Similarly, the number of isolates susceptible by both DD and E test were 10, 21, 19 and 25 by ertapenem, doripenem, meropenem, and imipenem, respectively. Result was marginally discordant (p=0.07) in ertapenem. E strip and disc diffusion results for other carbapenems i.e., imipenem, meropenem, and doripenem, were observed highly discordant (p<0.0001). Then, we compared the result of the ertapenem disc diffusion test and E test result with the gold standard assay i.e., CNP and/or PCR [Table/Fig-6]. Ertapenem disc diffusion test was observed to have higher sensitivity than the ertapenem E test. However, the specificity and PPV of both the tests were observed to be almost the same. On comparison of ertapenem disc diffusion test result with CNP and/or PCR, true positive, true negative, false positive, false negative for CP-CRE were observed in 80.2%, 6.5%, 3.9%, and 9.2%, respectively [Table/Fig-7].
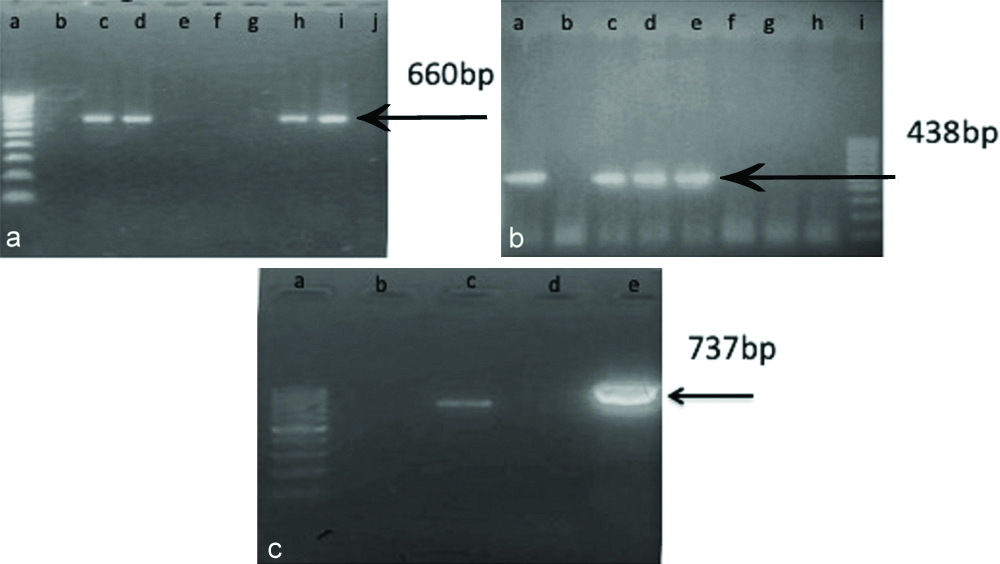
Agarose gel electrophoresis of PCR products from CRE isolates for blaNDM-1, blaOXA-48, blaIMP. a: The 660-bp amplicon present in lanes d, h and i; Lanes, e, f, g and j contain Negative sample for blaNDM-1; Lane, b contains Negative control; Lane, c, contain Positive control; Lane a, contain molecular marker (100bp). b: The 438-bp amplicon present in lanes c, d and e; Lanes, f, g and h contain Negative sample for blaOXA-48; Lane, b contains Negative control; Lane a, contain Positive control; Lane i, contain molecular marker (100bp). c: The 737-bp amplicon present in lane, e; Lane, d contain Negative sample for blaIMP; Lane, b contains Negative control; Lane, c, contain Positive control; Lane, a, contain molecular marker (100bp).
Comparative evaluation of the performance of various carbapenem disc and their combination for detection of carbapenem resistance.
| Disc diffusion | Sensitive | Resistant |
|---|
| Imipenem | 26 | 50 |
| Meropenem | 19 | 57 |
| Doripenem | 22 | 54 |
| Ertapenem | 12 | 64 |
| Imipenem+Meropenem | 17 | 59 |
| Imipenem+Doripenem | 20 | 56 |
| Meropenem+Doripenem | 16 | 60 |
| Imipenem+Meropenem+Doripenem | 29 | 47 |
| Imipenem/Meropenem/Doripenem | 15 | 61 |
Comparison of E test of imipenem, meropenem, doripenem, and ertapenem for detection of Carbapenem Resistance Enterobacteriaceae.
N.B: MIC breakpoints of Imipenem/Doripenem/Meropenem (susceptible, ≤1 μg /mL; intermediate, 2 μg/mL; resistant, ≥4 μg/mL) and for Ertapenem (susceptible, ≤0.5 μg /mL; intermediate, 1 μg/mL; resistant, ≥2 μg/mL).
Comparison of disc diffusion test with E test of individual carbapenem disc.
| Disc | DD E test S S | DD E test S R | DD E test R S | DD E test R R | p-value* |
|---|
| Imipenem | 25 | 01 | 32 | 18 | <0.0001 |
| Meropenem | 19 | 00 | 34 | 23 | <0.0001 |
| Doripenem | 21 | 01 | 36 | 18 | <0.0001 |
| Ertapenem | 10 | 02 | 09 | 55 | <0.07 |
NB: DD: Disc diffusion, R: Resistant, S: Sensitivity
*Result of either CNP or PCR was taken as gold standard while comparing the result of disc diffusion and E test using Chi-square test
Comparison of ertapenem disc diffusion test with ertapenem E test.
| Test type | Sensitivity | Specificity | PPV | NPV | p-value* |
|---|
| Ertapenem disc diffusion | 89.7% | 62.5% | 95.3% | 41.7% | 0.34 |
| Ertapenem E test | 79.4% | 62.5% | 94.7% | 26.3% | 0.12 |
NB: *Positive by either CNP or PCR was taken as gold standard while comparing the result of disc diffusion and E test using Chi-square test
Comparison of ertapenem disc diffusion test with CNP/PCR.
| Ertapenem DD | CNP/PCR |
|---|
| Sensitive | Resistant | Total |
|---|
| Sensitive | 5 (6.58%) | 7 (9.2%) | 12 (15.7%) |
| Resistant | 3 (3.95%) | 61 (80.2%) | 64 (84.2%) |
| Total | 8 (10.5%) | 68 (89.4 %) | 76 (100%) |
NB: DD: Disc diffusion
Discussion
The emergence of CRE is becoming a potential threat in patient care both in the hospital as well as community settings. There is a varying degree of expression of carbapenem resistance due to the presence of diverse amount of different carbapenemase, ESBL, and AmpC, non-enzymatic mechanisms like alteration in efflux pumps and mutation in the porin channels [15]. Carbapenemases are a group of hydrolytic enzymes that attack carbapenem drugs and neutralise it. They are usually carried in the mobile genetic elements such as plasmids or transposons. Several types and subtypes of these enzymes are present, depending upon their preferred substrate and molecular structure. Increased MIC may happen due to a combination of these mechanisms. In contrast, decreased MIC may happen due to the presence of isolated mechanisms, especially the non-enzymatic mechanisms like loss of porin channels. Correct identification of CRE is essential to provide appropriate therapy and follow the infection control protocols such as isolation and standard precaution for decrease its spread in the healthcare setting [16]. As per CDC 2015, the definition of CRE had included ertapenem with other carbapenems, thereby increasing the sensitivity of CRE detection [5]. However, there is very little literature comparing ertapenem disc and ertapenem E tests against most of the tests used for CRE detection as a screening tool [4,17].
Non-susceptibility of ertapenem primarily detects the beta-lactamases activity and/or other non-carbapenemase producing mechanisms [9,17]. Although some reports are published worldwide, very few reports have been documented from the clinical microbiology laboratory [10,15]. The disc diffusion and MIC determination are the two most common methods for phenotypic detection of carbapenem resistance. Ertapenem has been considered superior to imipenem and meropenem in terms of sensitivity for detecting carbapenem resistance by many studies [9,11,17]. Behera B et al., and Leavitt A et al., observed lower ertapenem MIC were susceptible to imipenem and meropenem due to ESBLs other than carbapenemases and loss of porin channels, Outer Membrane Porin K. pneumoniae-36kDa (OMPK36). Present study result was also observed in concordance with that which might be due to mechanisms other than carbapenemases production [10,17]. In the present study maximum concordance between the ertapenem disc diffusion test and the ertapenem E strip test was observed. This highlights the usefulness of ertapenem disc as a useful screening marker for detecting carbapenem resistance.
When ertapenem disc and ertapenem E strip tests were individually compared with CNP and/or PCR, the sensitivity and the NPV of the ertapenem disc diffusion test were observed to be higher than the ertapenem E strip test. However, the specificity and PPV were found the same as the E strip test. It may be due to the detection of non-carbapenemase based resistance mechanisms by the disc diffusion tests, which might have missed by the E strip tests due to low MIC levels. The ertapenem disc diffusion test detected 80% of the total CP-CRE. NonCP-CRE strains, in comparison to CP-CRE strains, are less virulent, less fit to the environment, hence less transmissible. There is limited literature available describing the prevalence of CP-CRE and nonCP-CRE separately [14]. Approximately, 3.95% of the isolates detected resistant by the ertapenem disc diffusion test were sensitive by CNP/PCR. It may be due to the detection of resistance by the mechanisms other than carbapenemase production such as AmpC/ESBL production or loss of porin channels, etc. Around 9.2% of the total isolates gave false-negative results, which may be due to the degradation of the drug. Present study result showed that the ertapenem disc diffusion test could detect both CP-CRE and nonCP-CRE isolates.
Limitation(s)
This study has several limitations. The number of isolates were less, and PCR was only done for limited carbapenemase-producing genes. The increased detection of CRE by ertapenem in comparison to other carbapenems due to non-carbapenemase mechanisms such as the production of AmpC, ESBLs, and alteration of porin channels could have been confirmed to establish the findings in the current study.
Conclusion(s)
In the current study, ertapenem disc diffusion test was observed to have less discordant result with the corresponding E test in comparison to other carbapenem discs i.e., imipenem, meropenem, and doripenem. It is able to detect both CP-CRE and nonCP-CRE organisms, which might be missed by the E test or CNP or PCR. Future studies with large sample size should be planned to evaluate the clinical and diagnostic significance of these results.
NB: DD: Disc diffusion, R: Resistant, S: Sensitivity
*Result of either CNP or PCR was taken as gold standard while comparing the result of disc diffusion and E test using Chi-square test
NB: *Positive by either CNP or PCR was taken as gold standard while comparing the result of disc diffusion and E test using Chi-square test
NB: DD: Disc diffusion