Salmonella Newport Gastroenteritis in an Adult- A Rare Case Report
Poornima Baby1, Aishwarya Babu2
1 Assistant Professor, Department of Microbiology, Amrita Institute of Medical Sciences and Research Centre, Kochi, Kerala, India.
2 Tutor, Department of Microbiology, Bharati Vidyapeeth Medical College Hospital and Research Centre, Pune, Maharashtra, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Poornima Baby, Assistant Professor, Department of Microbiology, Amrita Institute of Medical Sciences, Ponekkara P.O., Kochi-682024, Kerala, India.
E-mail: poornimasankar0709@gmail.com
Salmonella Newport is a major cause of food-borne infection which occurs due to consumption of contaminated food items. Stool sample from a suspected case of enteric fever was received in the Central Microbiology Laboratory of a tertiary care teaching hospital in Southern India. The bacterial isolate was identified on the basis of Gram Staining, cultural characteristics and biochemical reactions as Salmonella species. Agglutination for serotyping was done and it was found to be agglutinable by only Polyvalent O antiserum. For further speciation, the isolate was sent to the National Institute of Cholera and Enteric Diseases (NICED), Kolkata, West Bengal, India and was identified as Salmonella enterica serotype Newport. The patient responded well to ciprofloxacin therapy. As the diagnosis of Non-Typhoidal Salmonellosis (NTS) is often challenging, patients with suspected Salmonella infections are usually given empirical antibiotic therapy which can cause an increase in drug resistant NTS.
Acute gastroenteritis, Diarrhoea, Non-typhoidal salmonellosis
Case Report
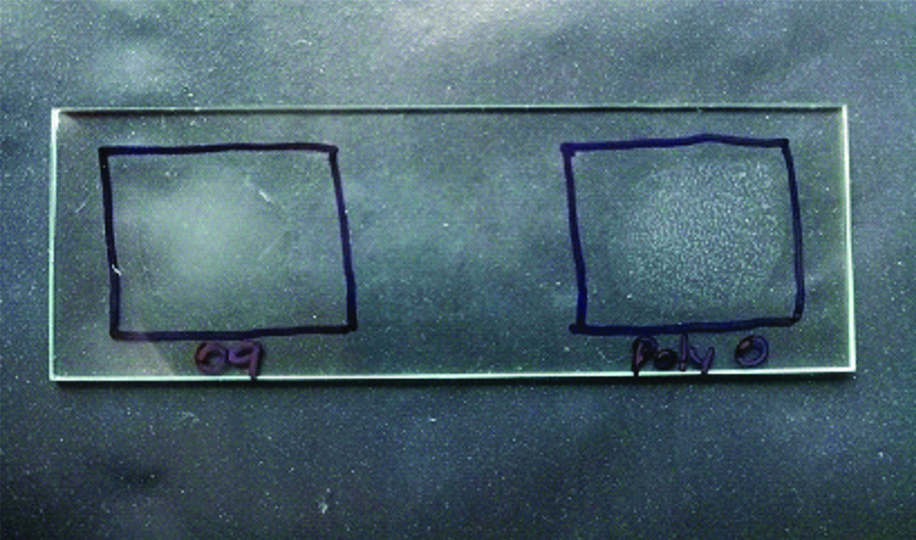
A 42-year-old lady came to the Medicine Outpatient Department (OPD) of a tertiary care teaching hospital in Southern India with complaints of fever and loose stools of two days duration. She gave a history of consumption of parottas (food item made of refined flour) and salads from a wayside restaurant three days prior to the onset of symptoms. There was no history of similar complaints in the other family members who had also consumed the same food. There was no history of contact with animals. As the case was initially thought to be one of acute gastroenteritis, an empirical treatment was initiated as per hospital policy and the patient was started on oral ciprofloxacin 5oo mg twice daily for seven days. Blood culture did not yield any growth. Blood routine and urine routine investigations were found to be within normal limits. Stool sample was received in the Microbiology Laboratory for culture and sensitivity. The wet film of the sample showed no pus cells, no red blood cells and no parasites. The routine culture and enrichment of the sample was done. It was incubated overnight aerobically at 37°C. The next day non-lactose fermenting colonies appeared on MacConkey Agar [Table/Fig-1]. The subculture from enrichment media after 18 hours of incubation also yielded similar colonies. The isolate was a gram negative, motile bacilli which was catalase positive, oxidase negative, nitrate reducing and glucose fermenting. Indole was not produced, citrate was utilised. Mannitol was fermented, triple sugar iron agar showed alkaline slant by acidic butt with hydrogen sulphide production and without gas [Table/Fig-2]. Lysine was decarboxylated. Ortho-nitrophenyl-β-D-Galactopyranoside (ONPG) test was negative. Agglutination test was positive only with Salmonella Polyvalent O antiserum [Table/Fig-3]. Agglutination test was negative for O9, O2, and O4. The antibiotics were tested according to Clinical and Laboratory Standards Institute (M100, 27th edition-2017) [1] and the organism was found susceptible to ampicillin, cotrimoxazole, ceftriaxone, ciprofloxacin and chloramphenicol. Since the isolate could not be serotyped using conventional methods, for further speciation the isolate was sent to NICED, Kolkata, West Bengal, India and was identified as Salmonella enterica serotype Newport. The patient was given oral ciprofloxacin 500 mg twice daily for seven days and when she came for follow-up her symptoms were found to have improved and she was advised to stop the antibiotics.
Non-lactose fermenting colonies on MacConkey agar.
Biochemical reactions of the isolate.

Agglutination tests: positive with Polyvalent O antisera; Negative with O9 antisera.
Discussion
Salmonellosis is a major public health problem worldwide [2]. Salmonella infections in humans often result from the ingestion of contaminated food such as poultry, beef, pork, milk and seafood [3]. Direct contact with animals also results in transmission of Salmonella to humans [4]. Infections with non-typhoid Salmonella enterica serovars is a problem of global concern [5]. The global burden of NTS gastroenteritis is approximately 94 million cases and the deaths amount to 155,000 yearly [6]. They are associated with high morbidity and mortality similar to the typhoidal Salmonellae. Salmonella enteritidis, Salmonella Newport, Salmonella typhimurium are the most common serotypes frequently isolated. The major cause of food-borne infections among NTS is Salmonella Newport. S. Newport outbreaks have been associated with consumption of contaminated dairy, poultry, eggs, seafood, meat, fresh fruits and vegetables like tomatoes, alfalfa sprouts, cantaloupes and many other food products [7]. In the present case, the source of infection in the patient would have been the salads and other food items that she had consumed from the restaurant. Salmonellosis in humans is a type of self-limiting diarrhoea not warranting any antimicrobial therapy. Rarely, they might need prompt chemotherapy to prevent life threatening systemic infections [8]. In recent years, NTS are gaining eminence, but they are rarely being reported in India and the emergence of antimicrobial resistance among them is at present a serious concern [9,10].
The NTS like S. Newport affect both immunocompetent as well as immunocompromised individuals. Immunocompromised individuals including those with malignancy, human immunodeficiency virus, diabetes and those receiving corticosteroid therapy or immunotherapy are at an increased risk of acquiring the infection. There have been many reported cases of S. Newport in humans, with a wide array of clinical spectrum such as diarrhoea, ileocecal lymphadenitis, chest wall abscess, pyosalpinx, osteomyelitis, endocarditis, meningitis, splenic abscess, septicaemia and bacteremia [11,12]. In this case, the patient presented with diarrhoea of two days duration.
The identification of Salmonella is relatively easy in the microbiology laboratory and is generally done by a combination of biochemical and antigenic tests. Confirmation of the isolates is done by agglutination using corresponding anti-sera. Commercial identification systems make use of different principles like latex agglutination, Enzyme Immunoassay (EIA) and Enzyme Linked Immunosorbent Assay (ELISA). Rapid identification and sensitivity methods are also available which include real time Polymerase Chain Reaction (PCR), Pulse Field Gel Electrophoresis (PFGE), Multi Locus Sequence Typing (MLST), Multiple-Locus Variable-Number Tandem Repeat Analysis (MVLA), Single Nucleotide Polymorphism (SNP) assays, Whole Genome Sequencing (WGS) and Matrix Assisted Laser Desorption Ionisation Time-of-Flight (MALDI-TOF) Mass Spectrometry. Though these tests are available, they are performed only at reference laboratories and for routine diagnostics the conventional methods of testing are preferred [13,14].
Antibiotic resistance among NTS has been reported to most classes of antibiotics. They are especially known to produce Extended Spectrum Beta-Lactamases (ESBL) and cephalosporinases. The resistance to quinolones and fluoroquinolones have also increased recently. This trend started developing post the introduction of these drugs for the treatment of similar infections. Salmonella serotype Typhimurium DT104, being resistant to at least five antimicrobials, including ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline, has caused severe mortality in animals and humans worldwide. In addition to the resistance to five drugs found in Salmonella serotype Typhimurium DT104, the novel Salmonella serotype Newport, called the serotype Newport MDR-AmpC (Multidrug Resistance- Ampicillinase C), is also resistant to amoxicillin-clavulanic acid, cephalothin, cefoxitin, and ceftiofur and shows decreased susceptibility to ceftriaxone. Some serotypes of the Newport MDR-AmpC strains, are also resistant to gentamicin, kanamycin, and trimethoprim-sulfamethoxazole [10].
As Salmonella gastroenteritis is most often a self-limiting disease, antibiotics are currently recommended only for patients with severe disease who are at high risk for invasive disease; occasionally fluid and electrolyte imbalance may need correction. Antibiotics should be avoided in uncomplicated acute gastroenteritis as they do not seem to shorten the duration of symptoms and they might prolong the duration of convalescent carriage. Their unwarranted use can also promote the emergence of drug resistant strains [15].
Conclusion(s)
The worldwide emergence of MDR phenotypes among Salmonella serotypes, especially Salmonella serotype Newport and Salmonella serotype Typhimurium is of increasing concern. This case of Salmonella Newport is an eye-opener as it enunciates the importance of judicious use of antibiotics in NTS. Since the isolate was not a MDR organism the patient improved with empirical ciprofloxacin, but this conventional practice of initiating empirical antibiotics in most cases will only accelerate the emergence of antimicrobial resistant strains of the organisms.
Author Declaration:
Financial or Other Competing Interests: None
Was informed consent obtained from the subjects involved in the study? No
For any images presented appropriate consent has been obtained from the subjects. NA
Plagiarism Checking Methods: [Jain H et al.]
Plagiarism X-checker: Jul 18, 2020
Manual Googling: Aug 21, 2020
iThenticate Software: Oct 28, 2020 (2%)
[1]. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017 [Google Scholar]
[2]. Kumar Y, Gupta N, Vaish VB, Gupta S, Distribution trends & antibiogram pattern of Salmonella enterica serovars Newport in IndiaThe Indian Journal of Medical Research 2016 144(1):82-86.10.4103/0971-5916.19329327834330 [Google Scholar] [CrossRef] [PubMed]
[3]. Gomez TM, Motarjemi Y, Miyagawa S, Kaferstein FK, Stohr K, Foodborne salmonellosisWorld Health Stat 1997 50:81-89. [Google Scholar]
[4]. Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Ceftriaxone- resistant Salmonella infection acquired by a child from cattleN Engl J Med 2000 342:1242-49.10.1056/NEJM20000427342170310781620 [Google Scholar] [CrossRef] [PubMed]
[5]. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Food-related illness and death in the United StatesEmerg Infect Dis 1999 5:607-25.10.3201/eid0505.99050210511517 [Google Scholar] [CrossRef] [PubMed]
[6]. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, The global burden of nontyphoidal Salmonella gastroenteritisClin Infect Dis 2010 50(6):88210.1086/65073320158401 [Google Scholar] [CrossRef] [PubMed]
[7]. Zhao S, Qaiyumi S, Friedman S, Singh R, Foley SL, White DG, Characterization of Salmonella enterica serotype Newport isolated from humans and food animalsJ Clin Microbiol 2003 41(12):5366-71.10.1128/JCM.41.12.5366-5371.200314662912 [Google Scholar] [CrossRef] [PubMed]
[8]. Travers K, Barza M, Morbidity of infections caused by antimicrobial-resistant bacteriaClin Infect Dis 2002 34(Suppl. 3):S131-S134.10.1086/34025111988884 [Google Scholar] [CrossRef] [PubMed]
[9]. Shahane V, Muley V, Kagal A, Bharadwaj R, Nontyphoid Salmonellosis: Emerging infection in Pune?Indian J Med Microbiol 2007 25:173-74.0.1016/S0255-0857(21)02186-1 [Google Scholar] [CrossRef]
[10]. Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, Molyneux E, Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in MalawiClin Infect Dis 2008 46:963-69.10.1086/52914618444810 [Google Scholar] [CrossRef] [PubMed]
[11]. Tappe D, Müller A, Langen HJ, Frosch M, Stich A, Isolation of Salmonella enterica serotype Newport from a partly ruptured splenic abscess in a traveler returning from ZanzibarJ Clin Microbiol 2007 45:3115-17.10.1128/JCM.00844-0717634302 [Google Scholar] [CrossRef] [PubMed]
[12]. Singh A, Wilkins T, Schade RR, Salmonella Newport bacteremia in a 12-day-old infantJ Am Board Fam Med 2011 24:214-17.10.3122/jabfm.2011.02.10015421383223 [Google Scholar] [CrossRef] [PubMed]
[13]. Bell RL, Jarvis KG, Ottesen AR, McFarland MA, Brown EW, Recent and emerging innovations in Salmonella detection: A food and environmental perspectiveMicrob Biotechnol 2016 9(3):279-92.10.1111/1751-7915.1235927041363 [Google Scholar] [CrossRef] [PubMed]
[14]. Saltykova A, Wuyts V, Mattheus W, Bertrand S, Roosens NHC, Marchal K, Comparison of SNP-based subtyping workflows for bacterial isolates using WGS data, applied to Salmonella enterica serotype Typhimurium and serotype 1,4,[5],12:i:-PLoS ONE 2018 13(2):e019250410.1371/journal.pone.019250429408896 [Google Scholar] [CrossRef] [PubMed]
[15]. Lee LA, Puhr ND, Maloney K, Bean NH, Tauxe RV, Increase in antimicrobial resistant Salmonella infections in the United States, 1989-1990J Infect Dis 1994 17:128-33.10.1093/infdis/170.1.1288014487 [Google Scholar] [CrossRef] [PubMed]