Antimicrobial resistance is one of major problem that world has to deal with urgency and there is an utmost need of finding the everlasting solution. The Centre for Disease Control and Prevention estimated that more than 2.8 million antibiotic resistant infections has occurred in United States [1]. In India also, there is a crisis of bacterial drug resistance; a report has found a mortality of 58,000 babies due to infections caused by multiple drug resistant bacteria [2]. Bacteria has evolved several mechanisms to avoid the action of antibiotics such as decreasing the intracellular accumulation of antibiotics, production of antibiotic inactivating enzymes, mutations of ribosomal proteins [3], and epigenetic mode of bacterial drug resistance [4]. A recent study has shown the presence of multi and extremely drug resistant Acinetobacter spp isolated from hospital, which is truly a worrisome due to its mode of bacterial drug resistant mechanisms [5]. A study from past decade has shown a novel epigenetic mode of bacterial drug resistance mechanism as a major factor in genesis of microbial drug resistance [6]. Literature revealed that the presence or expression of the methyltransferase enzymes causes the methylation of 16S rRNA (ribosomal ribonucleic acid, where S (Svedberg)) leading to resistance to aminoglycosides in gram negative bacteria [6].
Co-production of different enzymes due to combined presence of different drug resistance causing genes is of a grave concern at the community level and in treating the patients. A study from Korean countries has found the co-production of qnrb and 16S rRNA methyltransferase in multidrug resistant enterobacteriaceae [7]. Al Sheik YA et al., has found the co-expression of the 16S rRNA methylase pre-dominantly armA and rmtB genes along with major β-lactamase genes [8]. Along with the β-lactamase genes, studies found co-expression of 16S rRNA methylase along with the carbapanemase which is of a grave concern. The carbapenem is broad spectrum, effective on both gram positive and gram negative bacteria, which is the last resort in treating the patients resistant to several beta-lactam drugs [9]. A very few studies found the co-expression of genes involved in resistance to aminoglycosides and carbapenems.
At the tertiary centre presence of multidrug resistant bacteria was found including resistance to carbapenem and aminoglycosides. Detection of different mechanisms of bacterial mode of drug resistance at the molecular level helps in the proper assessment and choosing the appropriate antibiotic at the appropriate time that will prevent the spread of bacterial drug resistance, thereby reducing the morbidity and mortality. The present study aimed for the detection of genes related to the methylation mode of bacterial drug resistant mechanism including armA, rmtA, rmtB, rmtC and rmtD as well as genes OXA-48, VIM and KPC in the production of carbapenemase, also aimed to identify the co-expression of these genes in the gram negative bacterial isolates.
Materials and Methods
Bacterial Isolates and Antibiotic Sensitivity Testing
The prospective study was conducted at the Department of Microbiology, Kamineni Academy of Medical Sciences and Research Centre, Hyderabad, Telangana, India. The study was conducted from April 2018 to August 2019. A total of 200 gram negative bacteria isolated from different specimens [Table/Fig-1] were received at Department of Microbiology, from different departments and sections of the hospital for the bacterial culture. These isolates were subjected to antibiotic susceptibility testing using Kirby-Bauer method according to the latest Clinical and Laboratory Standards Institutes (CLSI) guidelines [10]. The antibiotics used were amikacin, gentamicin, ciprofloxacin, ofloxacin, cefazolin, ceftazidime, ceftriaxone, cefaperzone sulbactum, netilmycin, piperacillin tazobactum, imipenem and meropenem [10].
List of clinical specimens from which different gram negative bacteria were isolated for the study.
| Specimen | Total number of bacterial isolates |
|---|
| Bile fluid | 1 (0.5%) |
| Blood | 12 (6%) |
| ET secretions | 40 (20%) |
| Pus | 13 (6.5%) |
| Sputum | 15 (7.5%) |
| Tissue | 19 (9.5%) |
| Urine | 90 (45%) |
| Wound swab | 10 (5%) |
| Total | 200 |
ET: Endotracheal aspirate
DNA Extraction
The multidrug resistant organisms were subjected to DNA extraction. The bacterial DNA was isolated using a DNA isolation kit (Hi PurATM, Bacterial genomic DNA purification kit, Hi Media, India) as per the instructions of the manufacturer. Quality of the DNA was assessed using 1.0% Agarose Gel Electrophoresis (AGE) and the isolated DNA was stored at -20°C [11].
A 16S rRNA Methyltransferase Gene Screening
The PCR was been performed using specific primers to screen the presence armA, rmtA, rmtB, rmtC and rmtD has been mentioned in [Table/Fig-2] [12].
List of primer sequence of different 16S rRNA methyltransferase genes mentioned along with amplification product size.
| Gene | Sequences | Product |
|---|
| armA | F-5′ATGGAT AAGAATGATGTTGTTAAG 3′ | 774 bp |
| R-5′TTATTTCTGAAATCCACTAGTAATTA 3′ |
| rmtA | F-5′ACTGTGATGGGATACGCGTC 3′ | 315 bp |
| R-5′AGCGATATCCAACACACGATGG 3′ |
| rmtB | F-5′ATGAACATCAACGATGCCCTC 3′ | 756 bp |
| R-TTATCCATTCTTTTTTATCAAGTATAT 3′ |
| rmtC | F-5′ATGAAAACCAACGATAATTATC 3′ | 846 bp |
| R-5′TTACAATCTCGATACGATAAAATAC 3′ |
| rmtD | F-5′ATGAGCGAACTGAAGGAAAAACTGC 3′ | 744 bp |
| R-5′TCATTTTCGTTTCAGCACGTAAAACAG 3′ |
The PCR was been performed using PCR reaction mix, 50-100 ng of bacterial DNA and the total volume of 25 μL. The conditions used for the PCR were; initial denaturation was carried out at 93°C for 5 minutes, followed by 35 cycles of denaturation at 93°C for 1 min, annealing at 55°C for 1 minute, extension at 72°C for 1 minute and a final extension at 72°C for 10 minutes. The final PCR product was checked in 1.5% agarose gel along with the 100 bp ladder.
Carbapenemase Genes Screening
The organisms resistant to imipenem and meropenem organisms were screened for the presence of carbapenemase genes. PCR was carried out in a total volume of 25 μL PCR reaction mixture, using specific primers including-
KPC-F-TGTTGCTGAAGGAGTTGGGC, R-ACGACGGCATAGTCAT TTGC.
OXA-48-F-AACGGGCGAACCAAGCATTTT, R-TGAGCAC-TTCTTTGTGATGGCT.
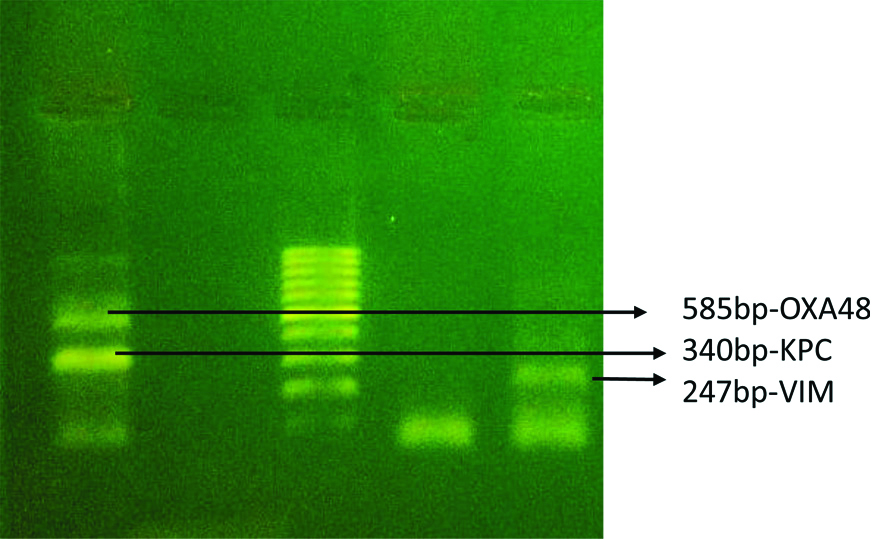
VIM-F-CGCGGAGATTGARAAGCAA, R-CGCAGCACCRGGATAGAARA (R=A or G) [12]. The multiplex PCR has been used for the detection of KPC, VIM and OXA-48. According to the instructions of Mlynarcik P et al., the PCR has been carried out in thermocycler (Takara, Japan) [13]. The amplification product obtained were of the expected size KPC-340 bp, VIM-247 bp and OXA-48 585 bp, has been checked using 1.5% agarose gel electrophoresis and 100 bp ladder as a marker in gel doc (Aga MIDI transilluminator).
Statistical Analysis
Data was compiled and percentage was calculated using MS-Excel.
Results
A total of 200 cases studied, including 68 females and 132 males. The mean age of the patients was 50.9 years (range min-3 years- max-95 years). The bacteria isolated were E.coli in 85 cases, Klebsiella pneumoniae in 55 cases, Pseudomonas aeruginosa in 40 cases and Acinetobacter Sp. were isolated in 20 cases. The antibiotic sensitivity pattern has been tabulated in [Table/Fig-3]. The aminoglycoside and carbapenem combined resistant organisms were isolated in 48 (24%) isolates. The armA gene was detected in 16 (13.2%) [Table/Fig-4a] isolates, rmtB detected in 15 (12.3%) [Table/Fig-4b], rmtC in 10 (8.2%) [Table/Fig-4c] and rmtD in 13 (10.74%) isolates [Table/Fig-4d,5]. No amplification of rmtA was noted. The carbapenemase genes were screened in carbapenem resistant isolates and found the presence of VIM in 20 (40%) of isolates, OXA-48 in 25 (50%) isolates and KPC is detected in 13 (26%) bacterial isolates [Table/Fig-5,6]. The co-expression of the 16S rRNA and carbapenemase genes were screened in multidrug resistant organisms mainly resistant to aminoglycosides and carbapenems, out of total 48 resistant organisms the co-expression of the genes were present in 9 (19%) isolates. The co-expression of different combination of methyltransferase and carbapenemase genes has been given in [Table/Fig-7].
Overall antibiotic sensitivity pattern of gram negative bacteria isolated from different specimens.
| Antibiotics | Resistance (cases) | Sensitive (cases) |
|---|
| Ciprofloxacin | 124 (62%) | 76 (38%) |
| Ofloxacin | 113 (57%) | 87 (43%) |
| Gentamicin | 121 (61%) | 79 (39%) |
| Amikacin | 69 (35%) | 131 (65%) |
| Netilimycin | 72 (36%) | 128 (64%) |
| Ceferperazone/Sulbactum | 69 (35%) | 131 (65%) |
| Pipercillin/tazobactum | 69 (35%) | 131 (65%) |
| Imipenem | 50 (25%) | 150 (75%) |
| Meropenem | 50 (25%) | 150 (75%) |
| Ceftazidime | 100 (50%) | 100 (50%) |
| Ceftriaxone | 95 (47%) | 105 (53%) |
| Cefataxime | 95 (47%) | 105 (53%) |
| Colistin | Nil | 100% |
Antibiotics susceptibility test done using Kirby-Baeur method according to the latest CLSI guidelines [10]
Detection of different 16S rRNA methyltransferase genes in multidrug resistant bacteria.
a.) It shows amplification of armA gene-774 bp and rmtD-744 bp.
b.) It shows amplification of rmtB gene-756 bp.
c.) It shows amplification of rmtC. gene-846 bp.
d.) It shows amplification of rmtC gene-846 bp, rmt-b-756 bp and armA-774 bp
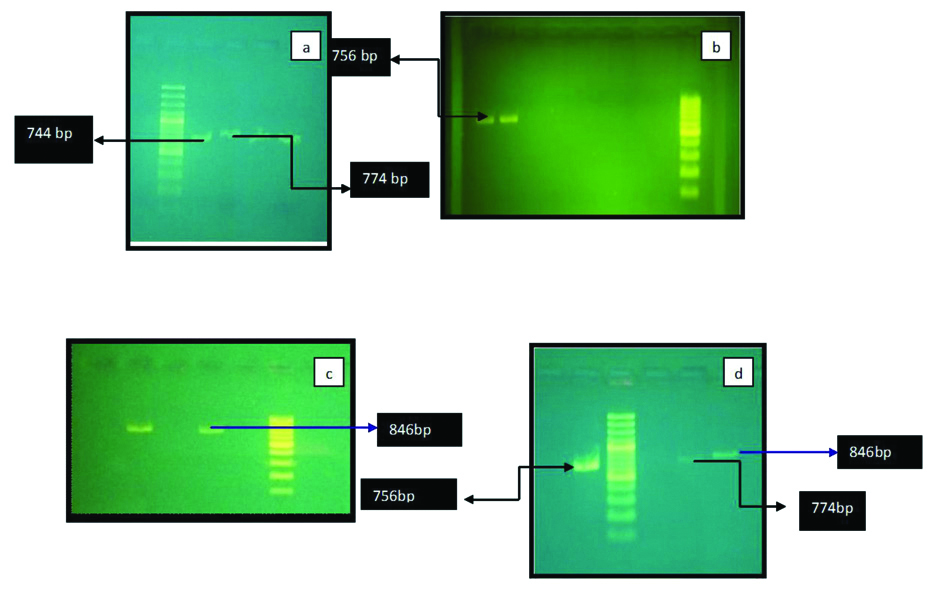
Distribution of bacterial drug resistance genes of 16S rRNA methyltransferase and carbapenmase genes in bacterial isolates.
| Bacterial isolates | 16S rRNA methyltransferase genes | Carbapenemse genes |
|---|
| Aminoglycoside resistance- (121 cases) 61% of total organisms | Carbapenem resistance-(50 cases) 25% of total organisms |
|---|
| armA | rmtB | rmtC | rmth | VIM | OXA48 | KPC |
|---|
| E.coli | 3 | 5 | 3 | 6 | 9 | 11 | 4 |
| Acinetobacter Spp. | 7 | 1 | 2 | 2 | 0 | 2 | 0 |
| Klebsiella Spp. | 4 | 7 | 4 | 3 | 6 | 10 | 6 |
| Pseudomonas Spp. | 2 | 2 | 1 | 2 | 5 | 2 | 3 |
| Total | 16 (13.2%) | 15 (12.3%) | 10 cases (8.2%) | 13 (10.7%) | 20 (40%) | 25 cases (50%) | 13 cases (26%) |
1. armA, rmtB, rmtC, rmtD- 16S rRNA methyltransferase genes
2. VIM, OXA-48, KPC- Carbapenemase genes
Detection of carbapenemase genes in multidrug resistant bacteria.
Co-expression of 16S rRNA methyltransferase and Carbapenem genes in gram negative bacteria resistance to both aminoglycosides and carbapenems (48 cases).
| Total number of organisms | Organisms | Genes |
|---|
| 1 | E.coli spp | rmtB, rmtD, OXA-48 |
| 2 | E.coli spp | rmtD, OXA 48 |
| 3 | E.coli spp | rmtB, rmtD, VIM |
| 4 | E.coli spp | rmtD, OXA-48 |
| 5 | Klebsiella spp | armA, rmtB, OXA-48, KPC |
| 6 | Klebsiella spp | armA, VIM, OXA-48, |
| 7 | Pseudomonas spp | rmtB, rmtD, VIM, OXA-48, KPC |
| 8 | Acinetobacter spp | rmtC, OXA-48 |
| 9 | Acinetobacter spp | rmtD, OXA-48 |
1. armA, rmtB, rmtC, rmtD- 16S rRNA methyltransferase genes.
2. VIM. OXA-48, KPC- Carbapenemase genes
Total number of organisms resistant to aminoglycoside and Carbapenem-48 Cases, Expression of both the genes-9 cases
Discussion
Multidrug resistant bacterium is one of the major factors which favour the spread of infection with high mortality and long stay in hospital. Screening of multiple genes of multiple drug resistant mechanisms helps us to deal the development of drug resistance in multiple ways. Bacteria evolved enumerable mechanisms to counter the action of antibiotics. The present study focused on the novel epigenetic mechanism of bacterial drug resistance especially involved in resistance to aminoglycosides. As bacteria are known to be the master of expressing different mechanisms to antibiotics action, the present study also evaluated the expression of the carbapenemase genes known to counter the action of carbapenem drugs having broad spectrum in action. We found 60% of carbapenem resistant organisms carrying the carbapenemase genes. To be called as multidrug resistant bacteria, the bacteria has to express different mechanisms through the activation of different genes leading to expression of different enzymes and act as one to inactive different antibiotics. To prove this point we evaluated the combined presence of 16S rRNA and carbapenem genes and found 19% of multidrug resistant organisms expressing both these genes.
The present study is the largest bacterial study to evaluate the co-expression of methyltransferase and carbapenem genes. Present study was in concordance with the study conducted by Hong SB et al., from South Korea, showed the co-existence of OXA-23 and the armA gene in multidrug resistant Acinetobacter species [14]. Similarly, a study by Lim J et al., has found the co-expression of the both OXA-23 and armA in more than 80% of cases [15]. This study also found the co-expression of the largely the OXA-48 gene has been seen expressed along with different methyltransferase genes. A study from the Poland echoed results finding the combination of epigenetic and carbapenem enzymatic action leading to drug resistance in Klebsiella species [16]. Absence of rmtA has been noticed in the present study, similar to the studies conducted by Hidalgo L et al., and Rehman M et al., [17,18]. These above mentioned studies found the absence of rmtA gene, and according to Mohanam L and Menon T [19], rmtA and rmtD have been frequently reported in European and American Countries contrary to these studies a study conducted by Upadhyay S et al., found the presence of rmtA in Acinetobacter spp [20]. Co-production of different enzymes and combined action of different mechanisms leads to the global dissemination of drug resistance, which has been emphasised through the present study and it is re-emphasised by Lee CS et al., who found the expression of NDM with methyltransferase rmtF found in a region of Minnesota [21]. The present study focused and studied the presence of wide range of methyltransferase and carbapenemase genes in multidrug resistant bacteria and for the first time to our knowledge found the combined combination of different methyltransferase genes associated with different carbapenemase genes. Individual screening for the genes involved in single drug resistant mechanisms may leave a void in fight against of global spread of drug resistant bacteria.
Limitation(s)
The present study has its own limitations as the drug resistance through minimum inhibitory concentration was not evaluated due to the limited sources, as well as epidemiological aspects was not focused.
Conclusion(s)
The present study found the presence of different methylase genes and high expression of carbapenem genes at individual level, and combined expression of these genes by bacteria may indicate to be utmost care to select the appropriate drug and emphasise the need of antibiotic stewardship leading to the prevention of global dissemination of multidrug resistant bacteria.
Antibiotics susceptibility test done using Kirby-Baeur method according to the latest CLSI guidelines [10]
1. armA, rmtB, rmtC, rmtD- 16S rRNA methyltransferase genes
2. VIM, OXA-48, KPC- Carbapenemase genes
1. armA, rmtB, rmtC, rmtD- 16S rRNA methyltransferase genes.
2. VIM. OXA-48, KPC- Carbapenemase genes
Total number of organisms resistant to aminoglycoside and Carbapenem-48 Cases, Expression of both the genes-9 cases