Identification of Dengue Serotypes using a Single Serum Specimen Algorithm in a Tertiary Care Hospital, Alappuzha, Kerala, India
Parvathy Vijayamohana Das1, Balakrishnan Anukumar2, Sobha Balakrishnan3
1 Assistant Professor, Department of Microbiology, Government TD Medical College, Alappuzha, Kerala, India.
2 Scientist ‘E’, National Institute of Virology Kerala Unit, Alappuzha, Kerala, India.
3 Professor and Head, Department of Microbiology, Government TD Medical College, Alappuzha, Kerala, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Parvathy Vijayamohana Das, Assistant Professor, Department of Microbiology, Government TD Medical College, Alappuzha-688005, Kerala, India.
E-mail: parvathyvdas@gmail.com
Introduction
The geographic location of Alappuzha, a district in the South Indian state of Kerala, the distinct weather conditions and frequent natural calamities present a unique ecology that contributes to the prevalence of vector-borne diseases like dengue. Early dengue virus infection can be detected by using a combination of tests on a single serum specimen.
Aim
To identify the dengue virus serotypes among hospitalised patients in a South Indian teaching hospital in Alappuzha, Kerala, India.
Materials and Methods
Patient samples that tested positive for dengue non-structural protein-1 (NS1) antigen by ELISA were further evaluated for dengue virus RNA by real-time, multiplex reverse transcriptase Polymerase Chain Reaction (PCR) and the serotype was determined. Anonymised patient data was collected using a questionnaire as a data collection tool. The data was analysed for statistical significance.
Results
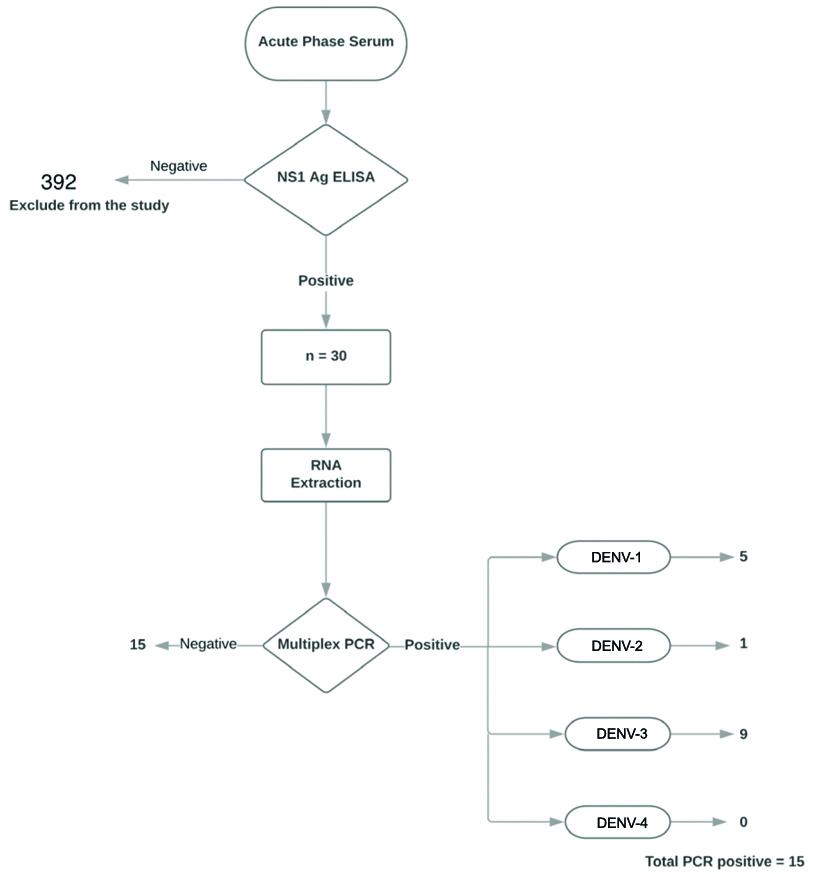
Among 422 non-duplicate patient serum samples received in the Department of Microbiology, in the year 2019, 30 were positive for dengue NS1 antigen by ELISA. Dengue viral RNA was detected in 50% of the samples (15/30). DENV-3 serotype was the most prevalent (nine) followed by DENV-1 (five) and DENV-2 (one). Common presentations of the patients were fever, headache, and myalgia. No statistically significant association was found between a PCR positive result and the presence of warning signs and thrombocytopenia.
Conclusion
DENV-3 was the most common serotype in the study population. Early dengue virus infection is associated with varied symptoms.
Arbovirus, Limited resource nation, Multiplex polymerase chain reaction, Real-time polymerase chain reaction, Reverse transcriptase polymerase chain reaction, Teaching hospital, Vector-borne disease
Introduction
Early diagnosis of dengue virus infection using sensitive and accurate laboratory tests is imperative for reducing mortality. Currently, assays that detect dengue-specific secretory NS1 antigen have gained popularity in the detection of early dengue virus infection [1,2]. Studies have shown a variable sensitivity for these assays (24-93%) in detecting dengue virus infection and a reduction of sensitivity in secondary dengue infections [3,4]. In secondary dengue infection, a rapid anamnestic rise in antibodies occurs. These antibodies sequester the NS1 antigen in immune complexes and thereby, reduce its detection through capture assays [5]. Nucleic acid detection methods are not widely used or are available in resource-limited countries like India. However, these assays are required for identifying the infecting serotype. The World Health Organisation (WHO) recommends their use for diagnosis of early dengue virus infection [6].
As per the guidelines put forth by the Directorate of National Vector-borne Disease Control Programme in India, a diagnosis of dengue fever is confirmed in a patient with clinical features of dengue by demonstration of viral nucleic acid, live virus, dengue NS1 antigen, or dengue IgM antibody in acute sera or by demonstration of IgG seroconversion or a four-fold increase in dengue IgG antibody titre in paired sera [7]. Demonstration of dengue IgG seroconversion or a four-fold increase in IgG titre in paired sera provides no useful information that influences early patient care. Moreover, collection of paired sera is often impractical and delays diagnosis. Most patients with dengue virus infection present acutely within a week of disease onset. A serum specimen collected during the acute phase is ideal for isolation of live virus or demonstration of viral nucleic acid or viral antigen. A previous study has demonstrated the efficacy of a single serum specimen in the diagnosis of dengue [8].
As of 2013, 19% of global cases of dengue infections were reported from India [9]. An increasing number of cases have been reported from the South Indian state of Kerala since 2006. In 2010, Kerala contributed to 9.2% of cases of dengue virus infections in India. All the four dengue serotypes (DENV-1,2,3 and 4) have been found to be prevalent in Kerala [10]. Some studies have reported an increased incidence of dengue hemorrhagic fever with DENV-2 infection [10-12]. The state of Kerala provides the ideal conditions for the study of neglected tropical diseases like dengue fever. Despite low per capita income, the health sector in Kerala has made outstanding developments by producing indices comparable at an international level. The high percentage of literacy and a strong public healthcare sector makes Kerala an ideal place for research on tropical infections [13]. In the previous year, 2018, Kerala was affected by severe floods. This natural calamity can potentially influence the epidemiology of these communicable diseases.
Alappuzha is a coastal district locked between a network of rivers, lakes and the Arabian sea. The abundance of water bodies around the district and the humid climate encourage vector-borne disease transmission. This study observed the clinical presentation in patients admitted with early dengue virus infection and attempted to identify the dengue viral serotypes.
Materials and Methods
This was a descriptive study conducted at a Tertiary Care Teaching Hospital in Alappuzha district of Kerala, India with the help of an attached virology lab. The study included all serum specimens that tested positive for dengue NS1 antigen by ELISA from January 2019 to September 2019. The study was begun after obtaining the approval of the Institutional Ethics Committee (IEC Certificate: EC43/2019 dated 09/05/2019).
A single serum specimen testing algorithm was followed for evaluating patients admitted in the hospital with a suspicion of dengue virus infection [7]. All serum specimens received in the microbiology lab with a clinician’s request for evaluation of dengue fever were tested for dengue NS1 antigen using commercially available sandwich ELISA. Sera that tested positive for dengue NS1 antigen were further subjected to a Multiplex, Real-Time, Reverse Transcriptase-Polymerase Chain Reaction (multiplex rRT-PCR). RNA isolation for the rRT-PCR was performed using QIAamp® Viral RNA Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Multiplex rRT-PCR assay was performed according to the protocol published by CDC (CDC DENV-1-4 rRT-PCR Multiplex Assay for Detection and Serotype Identification of Dengue Virus) [14]. This assay was performed using CFX96TM Real-Time system (Bio Rad Laboratories GmbH, Munich, Germany) [Table/Fig-1].
Multiplex real-time reverse transcriptase PCR assay.
1 Red line- Dengue serotype 3 positive control
2 Dark green line- Dengue serotype 2 positive sample
3 Dark green line- Dengue serotype 2 positive control
4 Blue line- Dengue serotype 1 positive control
5 Purple line- Dengue serotype 4 positive control
6 Blue line- Dengue serotype 1 positive sample
7 Red line- Dengue serotype 3 positive sample

A self-prepared, English questionnaire was used as a data collection tool to record clinical data of all patients that tested positive for dengue NS1 antigen [Annexure]. This questionnaire was designed based on the dengue case definition published by WHO [15]. Due importance was given to note the presence of warning signs as per CDC guidelines for dengue care management [16]. The questionnaire was face validated by internal medicine, community medicine and microbiology specialists and reliability of data collection was found appropriate. Anonymity of patient data was maintained throughout the study.
Statistical Analysis
All data were entered into Microsoft Excel. Unpaired proportions were compared by Fisher’s exact test. The p-value was calculated wherever required and value <0.05 was considered to be statistically significant.
Results
A total of 422 serum samples were received in the Department of Microbiology for diagnostic evaluation of clinically suspected dengue fever during the study period. This included 232 men and 190 women. Thirty (30/422) samples were found to be positive for dengue NS1 antigen by ELISA. Twenty one were men and nine were women, between the ages 18 and 70 years. Dengue viral RNA was detected in 15/30 (50%) of these patients who were positive for NS1 antigen by ELISA. The most common serotype isolated was DENV-3 (9/15) [Table/Fig-2]. Dengue viral RNA was detected in 6 women and 9 men out of 9 and 21 who tested positive for NS1 antigen, respectively.
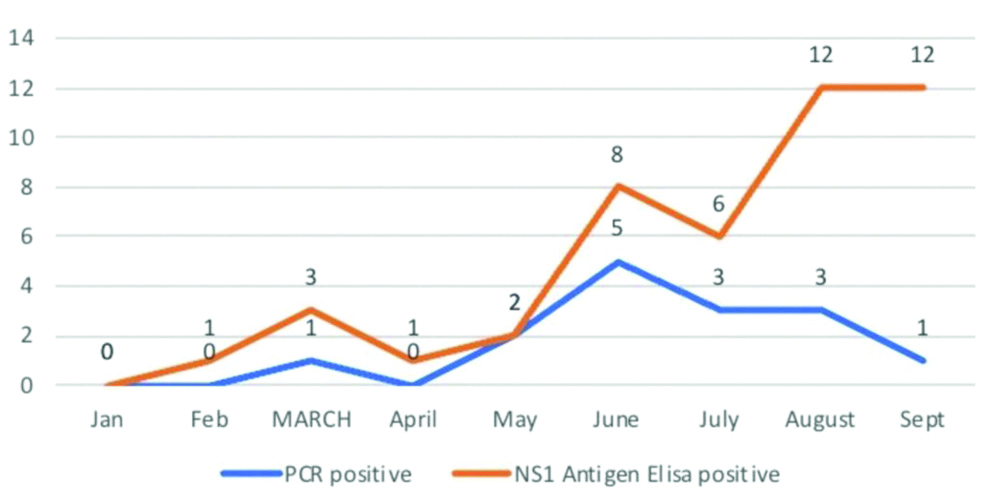
The maximum number of PCR positive cases was obtained in the month of June (5/8) [Table/Fig-3].
Single serum testing algorithm (original).
Month-wise distribution of results.
All 30 patients in the study population (NS1 antigen positive) had fever as the initial sign of illness. Out of the 15 who were PCR positive, two patients were afebrile at sample collection. The average duration of fever at sample collection was 3.9 days for PCR-positive patients (15) whereas it was 5.9 days for PCR-negative patients [15]. Other symptoms present in most patients included headache, myalgia and nausea. Retro-orbital pain was reported by two patients, who were both PCR negative. Among the 15 patients with a positive PCR result, only one patient had a clear history of previous dengue infection. Secondary dengue infection was not studied. Among the 15 patients with a positive PCR result, six had thrombocytopenia (platelet count <1,00,000). Among PCR negative patients, thrombocytopenia was seen only in eight patients. No statistical significance between the rRT-PCR test result and the presence of warning signs (p=0.47) or thrombocytopenia (p=0.75) was found [Table/Fig-4]. Though hepatomegaly was not observed in any of the patients, five patients (5/30) had moderate elevation of transaminases; dengue viral RNA was detected in two patients by PCR. Two patients had elevation of blood urea and serum creatinine; dengue viral RNA was detected in one patient.
Clinical symptoms and signs among the study subjects.
| PCR negative | PCR positive |
|---|
| Total | DENV-1 | DENV-2 | DENV-3 |
|---|
| Warning signs |
| Severe abdominal pain | 2 | 1 | | | 1 |
| Vomiting | 5 | 4 | 1 | | 3 |
| Mucosal bleeds | 2 | 1 | | 1 | |
| Laboratory investigations |
| Thrombocytopenia | 8 | 6 | 1 | 1 | 4 |
| Thrombocytopenia + leucopenia | 3 | 0 | | | |
| Elevated liver enzymes | 3 | 2 | | | 2 |
| Deranged renal function | 1 | 1 | | | 1 |
| Signs of severe dengue/ Organ involvement |
| Acute respiratory distress syndrome | 1 | 0 | | | |
| Acute coronary syndrome | 1 | 0 | | | 0 |
| Sinus tachycardia | 0 | 1 | | | 1 |
| Cerebrovascular event | 1 | 0 | | | |
One patient developed acute coronary artery syndrome and signs of severe dengue. This patient’s sample was negative by PCR. He was managed under intensive care with low-dose aspirin, repeated platelet monitoring, fresh frozen plasma infusion and cardiac monitoring. However, he developed Acute Respiratory Distress Syndrome (ARDS) during treatment and succumbed to it. Another patient had sinus tachycardia on ECG. He was managed with intensive cardiac monitoring and supportive therapy for dengue. This patient had a positive dengue PCR. His cardiac condition was stable during the hospitalisation and was discharged after supportive therapy for dengue. He was advised cardiology review after discharge.
Discussion
Dengue viral RNA was isolated from 50% of the samples tested by PCR. The most common serotype isolated was DENV-3. DENV-4 was not isolated from the samples. The present study happens to be the first attempt to identify the serotypes in a geographic location during a period of nine months within the year 2019. The study occurred in a Tertiary Care Government Teaching Hospital located in Alappuzha that caters to the entire population of Alappuzha. The absence of any other major hospitals or tertiary care centres in Alappuzha makes the study results an ideal representation of the community. Previous studies in India have reported the presence of all the four serotypes [Table/Fig-5,6] [10,17-24]. Among the studies from Kerala, one study included 12 serum samples from Alappuzha, of which only one sample yielded a positive PCR result. None of these studies have provided an ideal representation of the prevalent serotypes in the community in Alappuzha.
Previous studies from Kerala on the prevalent dengue serotypes [10,17-19].
| Study, Year of Publication | Period | Districts (Number of PCR positive samples) | PCR positive | Year wise distribution of PCR positive samples | Most common serotype |
|---|
| Present study, 2020 | 2019 | Alappuzha (15) | 50% (15/30) | 2019 (15) | DENV-3 |
| Kumar NP et al., 2013 [10] | 2008-2010 | Kottayam (19)Kozhikode (5)Kasaragod (4)Alappuzha (1) | 32% (29/89) | 2008 (1)2009 (8)2010 (20) | DENV-2 |
| Anoop M et al., 2010 [17] | 2008 | Ernakulam (37) | 49.3% (37/75) | 2008 (37) | DENV-2 |
| Anoop M et al., 2012 [18] | 2009 | Thiruvananthapuram Medical college campus (39) | 51.3% (39/76) | April-May 2009 (39) | DENV-1 |
| Reddy MN et al., 2017 [19] | 2013-2015 June-August | Kasargod | 26/100 | 2013 (13)2014 (3)2015 (10) | DENV-1 |
Previous studies from India on the prevalent dengue serotypes [20-24].
| Study, Year of Publication | Period | State | PCR positive | Year wise distribution of PCR positive samples | Most common serotype |
|---|
| Gupta E et al., 2006 [20] | 2003-2005 | Delhi (27) | 27/85 (32%) | 2003 (8)2004 (2)2005 (17) | All four serotypes (2003)DENV-1 (2004)DENV-3 (2005) |
| Vinodkumar CS et al., 2013 [21]. | 2011-2012 | Karnataka | 42/72 (58%) | June 2011-March 2012 (42) | DENV-2 |
| Barde PV et al., 2015 [22] | 2012 | Chhattisgarh | 53/142 (37%) | August 2012 (53) | DENV-1 |
| | Madhya Pradesh | 62/105 (59%) | November 2012 (62) | DENV-1 |
| Sharmila PF et al., 2019 [23] | 2017 | Tamil Nadu | 68/147 (46%) | August-October 2017 (68) | DENV-4 |
| Racherla RG et al., 2018 [24] | 2017 | Andhra Pradesh | 60/96 (63%) | Add (60) after 2017 | DENV-2 |
A single specimen diagnostic algorithm was previously used by a study which concluded that when real-time, reverse transcriptase PCR and NS1 antigen by ELISA were combined with dengue IgM antibody detection in a single serum specimen, dengue virus infection can be accurately detected in more than 90% of cases [8]. The same study demonstrated that over 90% of dengue infections can be confirmed if a combination of three assays- dengue viral RNA PCR, dengue NS1 antigen ELISA and dengue IgM ELISA- were performed on a single serum specimen collected within 10 days of onset of fever. Therefore, serum specimens collected early on in the infection and tested with a combination of nucleic acid assays, NS1 antigen ELISA and dengue antibody IgM ELISA can provide meaningful results that can aid in clinical practice. This inference forms the basis for a single serum specimen testing algorithm. However, the aforementioned study was conducted on archived samples preserved over a period of five years.
Dengue virus infection is a significant cause of febrile illness in Kerala. Similar studies conducted previously were able to isolate the dengue viral RNA from 49.3% of cases with clinical suspicion of dengue [17]. In another similar study conducted in Central India, dengue viral RNA was detected in 89% of samples which were positive for NS1 antigen by ELISA [25]. In the present study, dengue viral RNA was isolated from 50% of the samples tested. Though co-infection with two serotypes have been reported by several studies, co-infection was not observed in this study [19,21,24,26]. In the present study, DENV-4 was absent in the population studied. Previous studies have also reported the absence of this serotype for consecutive years [27,28]. A previous study from Karnataka employing a similar methodology, recovered dengue viral RNA from 25% of the samples tested and did not isolate DENV-4 serotype [29].
Five Indian states (Punjab, Haryana, Gujarat, Rajasthan and Kerala) were recently studied for the variability of vector-virus interactions and the influence of climate [30]. This study demonstrated that the extrinsic incubation period of dengue virus in the mosquito vector is the lowest in Kerala. The same study also inferred that the high incidence of dengue fever in Kerala is due to the abundance of breeding grounds, suitable temperatures (23.5-30°C) and shorter incubation periods. Epidemiological studies have shown that a secondary infection with a different serotype can cause severe dengue and the presence of more than one serotype in a geographic location can increase the risk for the same [31]. The present study has shown the prevalence of more than one serotype within Alappuzha. DENV-2, which has been described as the most virulent of dengue virus serotypes, was also isolated in the study [32]. The presence of more than one serotype in Alappuzha suggests endemicity and increases the risk for co-infection and severe disease. The geographic location of Alappuzha and the abundance of the vectors contribute to the increased risk for outbreaks. Lessons learned from the epidemiological surveillance of dengue may be applied to other emerging arbovirus infections like chikungunya and Zika virus infection, as these share common vectors [33,34].
Limitation(s)
Association between serotypes and specific symptoms or signs could not be established due to small sample size. Low-viral RNA, sensitivity of NS1 ELISA and specificity of PCR could be the reason for absence of viral RNA in 50% of the samples. Phylogenetic assay was not performed due to the lack of funding. The study had to be abruptly stopped by the month of September due to the same reason.
Conclusion(s)
The results of the study provided a veracious and current picture of the dengue virus serotypes circulating in the community in Alappuzha, Kerala, India. Since India has diverse population and geographic conditions, such small-scale studies can provide a better picture about the distribution of dengue virus serotypes in the country. Continuous local monitoring of infectious agents provides a database to predict outbreaks. Virus levels in the blood drop drastically within a few days of onset of illness. Early sample collection is required to culture the virus from blood or to demonstrate viral nucleic acid.
Author Declaration:
Financial or Other Competing Interests: None
Was Ethics Committee Approval obtained for this study? Yes
Was informed consent obtained from the subjects involved in the study? NA
For any images presented appropriate consent has been obtained from the subjects. NA
Plagiarism Checking Methods: [Jain H et al.]
Plagiarism X-checker: May 06, 2020
Manual Googling: Jul 08, 2020
iThenticate Software: Sep 16, 2020 (7%)
[1]. Cdc.gov. Testing Guidance |Dengue | CDC [Internet]. Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Vector-Borne Diseases (DVBD); [reviewed 2019 May 3; cited 2020 January 10]. Available from: https://www.cdc.gov/dengue/healthcare-providers/testing/testing-guidance.html [Google Scholar]
[2]. Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M, Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infectionsJ Clin Microbiol 2002 40(2):376-81.10.1128/JCM.40.02.376-381.200211825945 [Google Scholar] [CrossRef] [PubMed]
[3]. Hermann LL, Thaisomboonsuk B, Poolpanichupatam Y, Jarman RG, Kalayanarooj S, Nisalak A, Evaluation of a dengue NS1 antigen detection assay sensitivity and specificity for the diagnosis of acute dengue virus infectionPLoS Negl Trop Dis 2014 8(10):e319310.1371/journal.pntd.000319325275493 [Google Scholar] [CrossRef] [PubMed]
[4]. Chaterji S, Allen JC, Chow A, Leo YS, Ooi EE, Evaluation of the NS1 rapid test and the WHO dengue classification schemes for use as bedside diagnosis of acute dengue fever in adultsAm J Trop Med Hyg 2011 84(2):224-28.10.4269/ajtmh.2011.10-031621292888 [Google Scholar] [CrossRef] [PubMed]
[5]. Muller DA, Depelsenaire ACI, Young PR, Clinical and laboratory diagnosis of dengue virus infectionJ Infect Dis 2017 215(suppl_2):S89-95.10.1093/infdis/jiw64928403441 [Google Scholar] [CrossRef] [PubMed]
[6]. WHO. Dengue diagnostics: Proceedings of a joint TDR/WHO and PDVI workshop. Geneva: World Health Organization on behalf of the Special Training Programme for Research and training in Tropical Diseases. 2004;6:01-96. Available from: https://www.who.int/tdr/publications/documents/dengue_diagnostics.pdf [Google Scholar]
[7]. National Guidelines for Clinical management of Dengue Fever. Directorate of National Vector Borne Diseases Control Programme, Dte General of Health Services, Ministry of Health & Family Welfare, Government of India; 2015 [Google Scholar]
[8]. Hunsperger EA, Muñoz-Jordán J, Beltran M, Colon C, Carrion J, Vazquez J, Performance of dengue diagnostic tests in a single-specimen diagnostic algorithm [published correction appears in J Infect Dis. 2017;215(11):1774]J Infect Dis 2016 214(6):836-44.10.1093/infdis/jiw10326984143 [Google Scholar] [CrossRef] [PubMed]
[9]. Mid Term Plan for prevention and control of Dengue and Chikungunya. Directorate of National Vector Borne Diseases Control Programme. New Delhi. 2011 [Google Scholar]
[10]. Kumar NP, Jayakumar PR, George K, Kamaraj T, Krishnamoorthy K, Sabesan S, Genetic characterization of dengue viruses prevalent in Kerala State, IndiaJ Med Microbiol 2013 62(Pt 4):545-52.10.1099/jmm.0.052696-023288429 [Google Scholar] [CrossRef] [PubMed]
[11]. Fried JR, Gibbons RV, Kalayanarooj S, Thomas SJ, Srikiatkhachorn A, Yoon IK, Serotype-specific differences in the risk of dengue hemorrhagic fever: An analysis of data collected in Bangkok, Thailand from 1994 to 2006PLoS Negl Trop Dis 2010 4(3):e61710.1371/journal.pntd.000061720209155 [Google Scholar] [CrossRef] [PubMed]
[12]. Balmaseda A, Hammond SN, Pérez L, Tellez Y, Saborío SI, Mercado JC, Serotype-specific differences in clinical manifestations of dengueAm J Trop Med Hyg 2006 74(3):449-56.10.4269/ajtmh.2006.74.44916525106 [Google Scholar] [CrossRef] [PubMed]
[13]. Kutty VR, Historical analysis of the development of health care facilities in Kerala State, IndiaHealth Policy Plan 2000 15(1):103-09.10.1093/heapol/15.1.10310731241 [Google Scholar] [CrossRef] [PubMed]
[14]. Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus [published correction appears in PLoS Negl Trop Dis. 2013;7(7)]PLoS Negl Trop Dis 2013 7(7):e231110.1371/journal.pntd.000231123875046 [Google Scholar] [CrossRef] [PubMed]
[15]. World Health Organization, Regional Office for South-East Asia. Comprehensive Guideline for Prevention and Control of Dengue and Dengue Haemorrhagic Fever. Revised and expanded edition. New Delhi: WHO Regional Office for South-East Asia; 2011. Pp. 212 [Google Scholar]
[16]. Centers for Disease Control and Prevention. Dengue Case Management [Internet]. CDC. Available from: http://www.cdc.gov/dengue/resources/DENGUE-clinician-guide_508.pdf [Google Scholar]
[17]. Manakkadan A, Issac A, Mathew T, Philip S, Kareem NA, Unnikrishnan R, Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South IndiaIndian J Exp Biol 2010 48(8):849-57. [Google Scholar]
[18]. Anoop M, Mathew AJ, Jayakumar B, Isaac A, Nair S, Abraham R, Complete genome sequencing and evolutionary analysis of dengue virus serotype 1 isolates from an outbreak in Kerala, South IndiaVirus Genes 2012 45(1):01-13.10.1007/s11262-012-0756-322729802 [Google Scholar] [CrossRef] [PubMed]
[19]. Reddy MN, Dungdung R, Valliyott L, Pilankatta R, Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013-2015 in northern Kerala, IndiaPeerJ 2017 5:e2970Published 2017 Mar 1410.7717/peerj.297028316881 [Google Scholar] [CrossRef] [PubMed]
[20]. Gupta E, Dar L, Kapoor G, Broor S, The changing epidemiology of dengue in Delhi, IndiaVirol J 2006 3:9210.1186/1743-422X-3-9217083743 [Google Scholar] [CrossRef] [PubMed]
[21]. Vinodkumar CS, Kalapannavar NK, Basavarajappa KG, Sanjay D, Gowli C, Nadia NG, Episode of coexisting infections with multiple dengue virus serotypes in central Karnataka, IndiaJ Infect Public Health 2013 6(4):302-06.10.1016/j.jiph.2013.01.00423806706 [Google Scholar] [CrossRef] [PubMed]
[22]. Barde PV, Kori BK, Shukla MK, Bharti PK, Chand G, Kumar G, Maiden outbreaks of dengue virus 1 genotype III in rural central IndiaEpidemiol Infect 2015 143(2):412-18.10.1017/S095026881400061224667083 [Google Scholar] [CrossRef] [PubMed]
[23]. Sharmila PF, Vanathy K, Rajamani B, Kaliaperumal V, Dhodapkar R, Emergence of dengue virus 4 as the predominant serotype during the outbreak of 2017 in South IndiaIndian J Med Microbiol 2019 37:393-400.10.4103/ijmm.IJMM_19_33832003339 [Google Scholar] [CrossRef] [PubMed]
[24]. Racherla RG, Pamireddy ML, Mohan A, Mudhigeti N, Mahalakshmi PA, Nallapireddy U, Co-circulation of four dengue serotypes at South Eastern Andhra Pradesh, India: A prospective studyIndian J Med Microbiol 2018 36:236-40.10.4103/ijmm.IJMM_18_10930084417 [Google Scholar] [CrossRef] [PubMed]
[25]. Barde PV, Shukla MK, Joshi P, Sahare L, Ukey MJ, Molecular studies on dengue viruses detected in patients from Central IndiaIndian J Med Microbiol 2019 37:12-18.10.4103/ijmm.IJMM_18_37731424004 [Google Scholar] [CrossRef] [PubMed]
[26]. Savargaonkar D, Sinha S, Srivastava B, Nagpal BN, Sinha A, Shamim A, An epidemiological study of dengue and its coinfections in DelhiInt J Infect Dis 2018 74:41-46.10.1016/j.ijid.2018.06.02030100535 [Google Scholar] [CrossRef] [PubMed]
[27]. Shrivastava S, Tiraki D, Diwan A, Lalwani SK, Modak M, Mishra AC, Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 seasonPLoS One 2018 13:e019267210.1371/journal.pone.019267229470509 [Google Scholar] [CrossRef] [PubMed]
[28]. Barde PV, Godbole S, Bharti PK, Chand G, Agarwal M, Singh N, Detection of dengue virus 4 from central IndiaIndian J Med Res 2012 136:491-94. [Google Scholar]
[29]. Manthalkar PS, Peerapur BV, Demographic and clinical profile of patients infected with dengue virus serotypes 1, 2, and 3 in North KarnatakaJ Nat Sc Biol Med 2019 10:144-48.10.4103/jnsbm.JNSBM_207_18 [Google Scholar] [CrossRef]
[30]. Mutheneni SR, Morse AP, Caminade C, Upadhyayula SM, Dengue burden in India: Recent trends and importance of climatic parametersEmerg Microbes Infect 2017 6(8):e7010.1038/emi.2017.5728790459 [Google Scholar] [CrossRef] [PubMed]
[31]. Guzmán MG, Kourí GP, Bravo J, Soler M, Vázquez S, Morier L, Dengue hemorrhagic fever in Cuba, 1981: A retrospective seroepidemiologic studyAm J Trop Med Hyg 1990 42(2):179-84.10.4269/ajtmh.1990.42.1792316788 [Google Scholar] [CrossRef] [PubMed]
[32]. Rico-Hesse R, Microevolution and virulence of dengue virusesAdv Virus Res 2003 59:315-41.10.1016/S0065-3527(03)59009-1 [Google Scholar] [CrossRef]
[33]. Matysiak A, Roess A, Interrelationship between climatic, ecologic, social, and cultural determinants affecting dengue emergence and transmission in puerto rico and their implications for zika responseJ Trop Med 2017 2017:01-14.10.1155/2017/894706728717366 [Google Scholar] [CrossRef] [PubMed]
[34]. Fauci AS, Morens DM, Zika virus in the Americas-Yet another arbovirus threatN Engl J Med 2016 374:601-04.10.1056/NEJMp160029726761185 [Google Scholar] [CrossRef] [PubMed]