Materials and Methods
The present study was a prospective study which was carried out during the period of two years (April 2016-March 2018), 270 samples were taken from different Departments at the Tlemcen University Hospital Centre (Algeria). The concerned hospitalised patients were from different wards who had been hospitalised for at least 48 hours (surgery department, nephrology department, urology department, maternity department, paediatrics department) and were diagnosed with the following pathologies: carcinoma (41), urinary tract disorders (63), surgical operations (87), chronic renal failure and urine catheter (13), diabetes mellitus (4), digestive tract disorders (32), and caesarean section (30). The majority of patients were on antibiotic therapy, particularly beta-lactam antibiotics and aminoglycosides before or during sample collection (urine, catheter, blood, wound or dialysis fluid). Patients whose clinical signs did not suggest an infection and those who came for an outpatient clinic were excluded from the study. The study has been approved by the University Ethics Committee of the SNV/STU faculty of the University of Tlemcen (APPROVAL NUMBER/ID: DM RM 12). Each sampled patient signed his or her informed consent before the study.
Identification
Genus identification was performed by conventional tests (Gram staining, Catalase test, Bile Esculin Agar (Oxoid, Ltd), growth on 6.5% Sodium chloride hyper saline broth and Haemolytic activity); species identification was obtained by the API20Strep system (Biomérieux, France). Genotypic identification of Enterococcus species was realised by searching for the ‘tuf’ gene [16], using the following primers: Tuf-F 5’-CCAATGCCACAAACACTCGT-3’ and Tuf-R: 5’-CCTGAACCAACACAGTACGT-3’. The chromosomal DNA was recovered by the “Boiling” method [17]; in Tris Ethylene diamine tetraacetic acid (TE) buffer (10X) at a temperature of 100°C for 20 minutes, then in ice to create a thermal shock, followed by centrifugation at 13000 rpm/10 minute. The PCR conditions were optimised as follows: a total mix of 50 μL containing: 0.5 μL (2.5 U/μL), Taq polymerase (Invitrogen), PCR buffer 5X, 4 μL dNTP (250 μM for each base), 1 μL of each primer (tuf-F and tuf-R), 1 μL of each DNA sample. The PCR program consisted of 30 cycles at 94°C/30 seconds, 51°C/1 minute, 72°C/1 minute 30 seconds, and finally a last extension at 72°C/10 minutes in a thermal cycler (BioRad), the migration of each PCR product was performed on 1% agarose gel for 30 minutes and visualisation under ultraviolet. All obtained amplified products corresponding to 803 bp were sequenced to validate their identification. The amplified fragments were sequenced and compared with sequences available deposited in the GenBank at the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) using the BLAST program [18].
Antibiotic Sensitivity Test
Antibiotic susceptibility of Enterococcus was tested using the Mueller-Hinton agar diffusion method (Oxoid, United Kingdom) as recommended by the Comité de l’Antibiogramme de la Société Française de Microbiologie (CASFM) guidelines (2018) [19]. Eighteen antibiotic disks were applied: Vancomycin (30 μg), Teicoplanin (30 μg), Imipenem (10 μg), Ampicillin (10 μg), Amoxicillin (30 μg), Cefalexin (30 μg), Cefoxitin (30 μg,), Gentamicin (10 μg), Tobramycin (10 μg), Amikacin (30 μg), Neomycin (30 UI), Ofloxacin (5 μg), Ciprofloxacin (5 μg), Rifampicin (5 μg), Erythromycin (15 μg), Tetracycline (30 μg), Clindamycin (2 μg), Fosfomycin (5 μg). Enterococcusfaecalis ATCC 29212 was used as a control strain. The MIC of the following eight antibiotics was determined by the broth microdilution method according to the CASFM recommendations (2018) [19]: ampicillin, amoxicillin clavulanic acid, imipenem, gentamicin, amikacin, ciprofloxacin, vancomycin, and teicoplanin. The E-test strips (Biomérieux, France) confirmed the MIC of the two glycopeptides tested (Vancomycin and Teicoplanin).
Detection of ‘Van’ Resistance Genes by PCR and Sequencing
Extraction of plasmid DNA: The DNA of bacteria with phenotypic resistance on disc diffusion (intermediate or complete) to vancomycin and teicoplanin was purified using the “Nucleospin Tissue” kit from Macherey Nagel according to the supplier’s instructions. The performed PCR involved two genes vanA (5’-GGGAAAACGACAACAATTGC-3’ and 5’-GTACAATGCGGGCCGTTA-3’) and vanB (5’-ACGGAATGGGGAAGCCGA-3’ and (5’-TGCACCCGATTTCGTTCGTTC-3’) [20] in a total volume of 25 μL containing: Taq polymerase 0.1 μL, Buffer 2.5 μL, 3 μL dNTP, 1.5 μL MgCl2 (25 mM), primer at 1.25 μL, EQ 14.4 μL. The PCR program consists of 35 cycles at 94°C/1 minute, 54°C/1 minute, 72°C/1 minute 30 seconds, and finally a last extension at 72°C/7 minutes. in a thermal cycler (BioRad), the migration of each PCR product was performed on 2% agarose gel for 40 minutes and visualisation under ultraviolet [21]. DNA sequencing was carried out by the Genoscreen laboratory (Lille, France) using the Sanger sequencing method (ABI3730) [22], the sequence reaction protocol was the Big Dye V3.1 terminator. The succession of nucleotide databases was analysed using the database available on the NCBI website (http://www.ncbi nlm.nih.gov).
STATISTICAL ANALYSIS
Statistical analysis was performed using SPSS software (IBM SPSS statistics 20). The probability <0.05 (p<0.05) was considered statistically significant.
Results
One hundred and eleven isolates assigned to the genus Enterococcus were isolated from 270 samples. For the rest of the samples, as soon as the presumptive characteristics indicated different species, identification was systematically abandoned. They were distributed in four enterococcal species: E. faecalis 59 (53%), E. faecium 47 (42%), E. avium 2 (1.8%) and E. durans 3 (2.7%). The highest isolates were obtained from different surgical site infections (49%) (abscess and suppurative wounds), this percentage decreases slightly to (40%) in urine samples, 10% were from blood cultures and one strain from dialysis fluid. [Table/Fig-1] represents the distribution of enterococcal species isolated from the different clinical samples. The patients were mainly adults (84%) (n=227) versus (16%) (n=43) of children. Among the adult population, the number of women (137) was greater than that of men (90); the age group of adults was between 17 and 80 years for men and between 15 and 74 years for women. With a mean age of 310.7 days, children in paediatric department were aged between 4 days and 13 years.
Distribution of Enterococcus according to the different clinical samples.
| Species | Urine | Blood | Surgical site | Dialysis fluid | Isolates |
|---|
| E. faecalis | 23 (51%) | 6 (55%) | 30 (55.5%) | 0 | 59 (53%) |
| E. faecium | 22 (49%) | 5 (45%) | 19 (35%) | 1 (100%) | 47 (42%) |
| E. avium | 0 | 0 | 2 (4%) | 0 | 2 (1.8%) |
| E. durans | 0 | 0 | 3 (6%) | 0 | 3 (2.7%) |
| Total | 45 (40%) | 11 (10%) | 54 (49%) | 1 (0.9%) | 111 (100%) |
The results of antibiotic susceptibility tests of the tested isolates have often revealed an ineffectiveness of some molecules. In fact, with the exception of imipenem, which was active on 83% of E. faecalis, all tested beta-lactams showed resistance rates ranging from 51% for ampicillin to 100% for cephalosporins [Table/Fig-2]. The situation remains relatively the same for E. faecium, which is characterised by a total resistance to cephalosporins, but this resistance is still more pronounced against imipenem. The activity of aminoglycosides on E. faecalis was different according to the tested molecules, and whereas these isolates developed high resistance rates to neomycin (100%) and amikacin (98%), they were more sensitive to gentamicin (56%) than to tobramycin (46%). The phenomenon of resistance has also considerably affected other molecules; thus, authors have recorded resistance rates in the order of 100% to clindamycin, and fosfomycin. On the other hand, fluoroquinolones appear to be relatively more active against E. faecalis (ofloxacin 36%, ciprofloxacin 30%), than against E. faecium (66% and 44%, respectively). By comparing the results between the two studied species, statistical analysis revealed that the two species seem to have the same antimicrobial resistance profile, except for E. faecium which presents a higher percentage to imipenem, ampicillin, tobramycin and ofloxacin (p<0.05) [Table/Fig-2].
Resistance profiles of the two species E. faecalis and E. faecium.
| Antibiotics | E. faecalis (n=59) | E. faecium (n=47) | Probability p |
|---|
| Resistance | % | Resistance | % |
|---|
| Vancomycin | 0 | 0 | 2 | 4 | 0.109 |
| Teicoplanin | 0 | 0 | 2 | 4 | 0.109 |
| Imipenem | 10 | 17 | 16 | 34 | 0.042 |
| Ampicillin | 30 | 51 | 35 | 74 | 0.013 |
| Amoxicillin | 33 | 56 | 33 | 70 | 0.131 |
| Cefalexin | 59 | 100 | 47 | 100 | / |
| Cefoxitin | 59 | 100 | 47 | 100 | / |
| Gentamycin | 26 | 44 | 25 | 53 | 0.350 |
| Tobramycin | 32 | 54 | 38 | 80 | 0.004 |
| Amikacin | 58 | 98 | 45 | 96 | 0.429 |
| Neomycin | 59 | 100 | 47 | 100 | / |
| Ofloxacin | 21 | 36 | 31 | 66 | 0.001 |
| Ciprofloxacin | 18 | 30 | 21 | 44 | 0.132 |
| Rifampicin | 45 | 76 | 37 | 79 | 0.764 |
| Erythromycin | 48 | 81 | 40 | 85 | 0.609 |
| Tetracyclin | 53 | 90 | 43 | 91 | 0.771 |
| Clindamycin | 59 | 100 | 47 | 100 | / |
| Fosfomycin | 59 | 100 | 47 | 100 | / |
In addition to the resistance confirmation, the results of the MICs allowed the determination of the current resistance level [Table/Fig-3]. This accordance with the susceptibility profiles led to a fluctuation in MICs sometimes exceeding 512 μg/mL. The combination of clavulanic acid with amoxicillin did not improve the activity of the latter, while imipenem was characterised by a relative efficiency with a resistance rate of only 23%. The resistance rate of amikacin was higher than gentamicin’s, and even though aminoglycosides don’t have remarkable effectiveness against Enterococci. Glycopeptides have generally conserved their efficiency on the tested isolates in this work, but we are still seeing the emergence of two E. faecium with MIC in the order of 256 μg/mL for vancomycin but this resistance level has decreased against Teicoplanin (48 and 64 μg/mL).
Minimum inhibitory concentrations (μg/mL) of Enterococci.
| AntibioticsMIC | VA | TEC | CIP | IPM | AMC | AMP | GEN | AMK |
|---|
| S≤ | R> | S≤ | R> | S≤ | R> | S≤ | R> | S≤ | R> | S≤ | R> | S≤ | R> | S≤ | R> |
|---|
| 4 | 4 | 2 | 2 | 4 | 4 | 4 | 8 | 4 | 8 | 4 | 8 | 128 | 128 |
|---|
| MIC <1 | 69 | 73 | 20 | 44 | 32 | 33 | 36 | 0 |
| 1≤MIC≤2 | 29 | 25 | 45 | 15 | 4 | 7 | 8 | 0 |
| 2<MIC<8 | 0 | 0 | 4 | 18 | 6 | 0 | 4 | 0 |
| 8≤MIC<64 | 0 | 1 | 4 | 13 | 16 | 14 | 6 | 4 |
| 64≤MIC<256 | 0 | 1 | 7 | 7 | 16 | 12 | 21 | 8 |
| MIC=256 | 0 | 0 | 10 | 2 | 8 | 4 | 16 | 6 |
| MIC≥512 | 2 | 0 | 10 | 1 | 18 | 30 | 09 | 82 |
VA: Vancomycin; TEC: Teicoplanin; CIP: Ciprofloxacin; IPM: Imipenem; AMC: Amoxicillin/Clavulanic acid; AMP: Ampicillin; GEN: Gentamycin; AMK: Amikacin; S: Susceptibility; R: Resistance
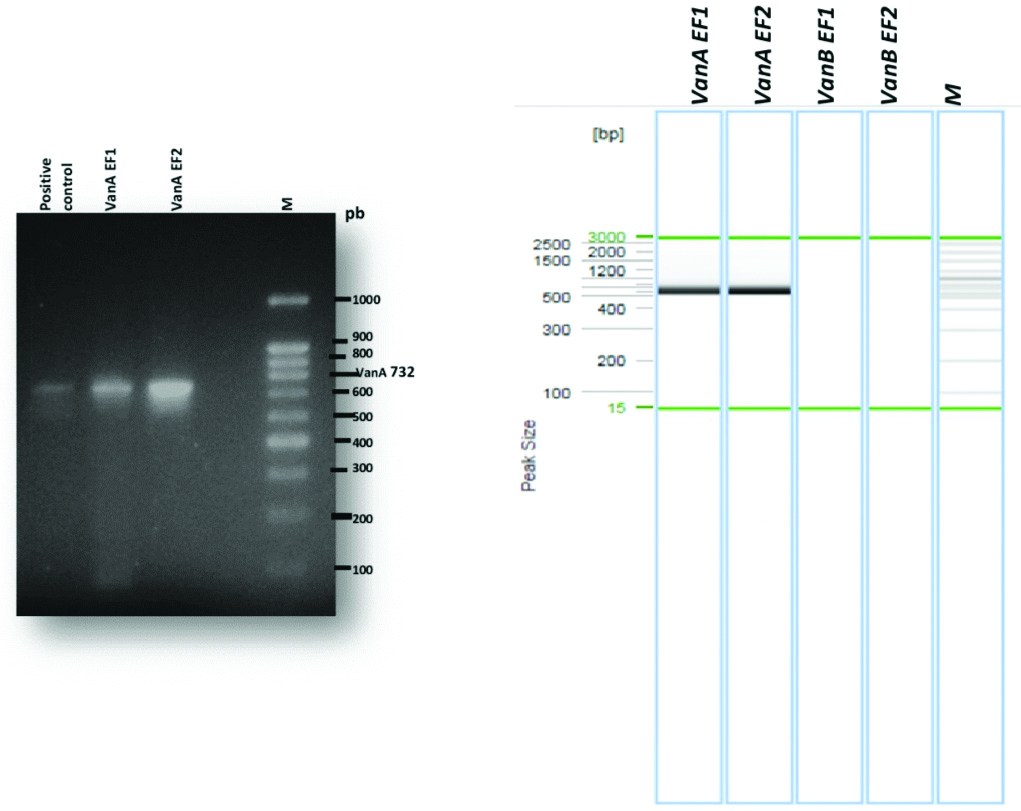
Genotypic characterisation of vancomycin resistance genes after migration by PCR showed the absence of the vanB gene and that the two-isolated VRE carry the vanA gene [Table/Fig-4]. The obtained sequence in this work corresponding to the vanA gene of the strain EF1 was submitted in the National Biotechnology Information Centre GenBank (http://www.ncbi.nlm.nih.gov/) under access number MH414912 (Enterococcusfaecium strain EF8RM D-alanine: D-lactate ligase (vanA) gene, partial cds). The two isolated Enterococcusfaecium carrying the vanA gene in this work came from two different samples, the first from dialysis fluid (EF1), in a 45-year-old patient with chronic renal failure and type II diabetes for 15 years, the haemodialysis frequency was three times per week via Arteriovenous Fistula (AVF), this patient had a history of catheter bacteremia and had received third-generation cephalosporin antibiotic therapy; the second sample (EF2) came from a blood culture of a patient who has undergone an emergency surgery for parietal sepsis and then transferred to the surgical department.
Analysis by DNA amplification of glycopeptide-resistant Enterococcus faecium.
1: Positive control; 2: EF1 (E. faecium); 3: EF2 (E. faecium), M: molecular size marker (100-bp DNA ladder; Invitrogen)
Discussion
This study focuses on the frequency and the resistance profile of isolated Enterococci in Algeria, using different samples from several departments of the Tlemcen University Hospital Centre, Algeria. Two predominant Enterococci species have been found: E. faecalis (53%), and E. faecium with 42%. This species distribution was comparable to a work realised in Turkey where E. faecalis was predominant over E. faecium isolates in hospitalised patients [23]. The results confirm the multidrug resistance state of Enterococci, which has never stopped to evolve, so after the appearance in the 1970’s of a high level resistance to ampicillin [24], the phenomenon has affected other classes of antibiotic molecules such as aminoglycosides, fluoroquinolones and glycopeptides, not even sparing linezolid, daptomycin and tigecycline, drugs considered as the ultimate defence against serious enterococcal infections, and successfully used in recent years to treat some infections [25,26].
The ampicillin resistance rate recorded in this work remains relatively similar to that reported in Cameroon and Ethiopia with rates of around 60% and 66.7%, respectively [27]. However, it is more important than the one reported by Komiyama EY et al., [28]. The resistance of Enterococci to beta-lactam antibiotics is generally due to two distinct mechanisms: an overproduction of Penicillin-Binding Protein 5 (PBP5) with a low affinity for beta-lactams or a synthesis of beta-lactamases, which appears much more pronounced in E. faecium than in E. faecalis [7]. In Turkey, a study showed a difference in reaction to ampicillin between E. faecalis and E. faecium, with resistance rates of 2% and 71%, respectively [23]. This resistance is expressed by the production of a plasmid PBP5 alone or in combination with punctual mutations, in particular PBP5 M485A with Ser insertion at position 466 in E. faecium [1].
The effect of ciprofloxacin against E. faecalis was relatively more significant, with a resistance rate of 30% [Table/Fig-2], which was still lower than that reported by Saeidi S et al., (43%) [29]. This resistance is often due to the active efflux system encoded by the emeA gene, which affects several molecules and is at the origin of the multidrug resistance phenomenon. In fact, in Enterococci, the use of reserpine, an active efflux pump inhibitor, significantly reduces the MICs of different molecules such as ciprofloxacin, gatifloxacin and levofloxacin [6]. The data recorded in this work confirm the state of multidrug resistance that affects the majority of tested isolates; in matter of fact, the extent of the phenomenon is such that one isolate (EF1) has shown resistance to all the tested antibiotics, and that on average eight isolates of E. faecium have been resistant to 11 antibiotics, which considerably limits the therapeutic treatments. This situation seems to reflect the care practices and the therapeutic choices specific to each service, also, according to an observance survey in the surgical service (Tlemcen hospital), the most commonly antibiotic used in 2017 is represented by cephalosporins (Cefazolin 35%, Cefotaxim 17%, and Ceftizoxim 14%) generating a selection pressure responsible of these resistance emergence [30].
The performed amplification reactions in this work in two E. faecium confirmed the presence of the vanA gene [Table/Fig-4], This result agrees with those reported respectively in Algeria, Morocco and Tunisia [31-33]. While another study carried out in Algeria, highlighted the presence of the VanC and Van C-1 gene in E. gallinarum [34,35]. The distribution of these resistance genes varies from country to country; therefore, the present results show the absence of the vanB gene in the tested isolates. While a study carried out in Turkey confirmed the absence of the vanB gene in E. faecium [23,36], another work carried out in South Africa reported a preponderance of the vanB, vanC1 and vanC2/3 genes in Enterococcus spp [21]. In fact, these two isolates have been resistant to gentamicin, which is in accordance with the work carried out in Cuba by Quinones D et al., [37], who reported high levels of resistance to this drug. This resistance has not spared beta-lactams such as ampicillin and imipenem which concords with the result of Celik S et al., [23]. Whereas the presence of the vanA gene is synonymous with resistance to vancomycin, some authors have identified a discordance between the genotype and phenotype by characterising vancomycin-sensitive Enterococci while hosting the vanA gene, the discrete presence of this genetic bearer might cause a vigilance decrease towards these isolates, allowing a silent diffusion of this gene [38].
In this study, resistance of two E. faecium seems to concern other antibiotic families, such as fluoroquinolones, which suggests the concomitant acquisition of several resistance mechanisms. These results seem to agree with those of Benammar S et al., on antibiotics such as amoxicillin, gentamycin and ciprofloxacin [31]. This agreement is not complete because it affects a wider range of antibiotics expressing total resistance to fosfomycin, and variable to erythromycin and tetracycline. These are the result of different rearrangements. Several authors have reported this genetic polymorphism. Actually, 25 vanA variants have been identified in 27 different countries, and though the variant containing the IS 1216 insertion sequence in the VanX-VanY and VanS-VanH pair remains the most predominant in the world, other variants have appeared in America and Europe with the IS 1251 sequence in the VanS and VanH region [39]. The vanA gene has also been found on a portion of the chromosome in an isolated strain of E. faecium in Germany, suggesting that the limit between the chromosome and the plasmid is ephemeral and that therefore, vancomycin resistance could be disseminated in both directions: vertical and horizontal [40], and allowing to other microorganisms to complete their genetic heritage by acquiring vancomycin resistance. This transfer could be intraspecific [41] as it can be interspecific by affecting species like E. faecalis [42,43] S.aureus, already found in the same hospital in Tlemcen [44] or Clostridium difficile [45]. This ensures both the sustainability and the dissemination of these genes, minimising the means of combating Enterococcus infections and thus, increasing the risk of therapeutic failure.
Limitation(s)
Permanent genetic rearrangements seem to be a hindrance to shedding light on the evolution of resistance. In order to understand how this type of strain circulates in Africa, it would be interesting to explore genetic polymorphism in order to establish a strategy that is both effective and adapted to each situation.
Conclusion(s)
This work confirmed the multidrug resistance status of both E. faecalis and E. faecium species affecting several families of antibiotics such as beta-lactamines, aminoglycosides, fluoroquinolones and glycopeptides. This phenomenon seems to be correlated to the antibiotics used specific to each department, leading to an increasing adaptation to these drugs that probably will not stop spreading.
Today, we are witnessing in Algeria the emergence of two new vancomycin-resistant E. faecium that constitute a potential reservoir of resistance genes such as the vanA gene that could easily access other species, limiting considerably molecules with stable and durable activity. It would be interesting to explore Clonal relatedness of Enterococci using Multilocus Sequence Typing (MLST), while tracking the circulation of other resistance genes.
VA: Vancomycin; TEC: Teicoplanin; CIP: Ciprofloxacin; IPM: Imipenem; AMC: Amoxicillin/Clavulanic acid; AMP: Ampicillin; GEN: Gentamycin; AMK: Amikacin; S: Susceptibility; R: Resistance