The excessive and inappropriate use of antibiotics has led to drastic increase in the prevalence of drug resistant bacteria in the world especially in the Indian population [1].
The microbial flora commonly present in dentulous patients and those with dentures are as follows: Actinobacillus actinomycetemcomitans, Bacteroides, Streptococci, Staphylococcus aureus, Lactobacilli, Enterobacteriaceae, Tetrads, Micrococci. Coccoid Gram-Positive Cocci, Gram-Negative Bacilla and Diplococci [2]. These bacteria which colonise the oral and the denture surfaces acquire antibiotic resistance due to frequent usage of antibiotics. These resistant bacteria pose a potential health hazard to the public and enigma to the clinicians while managing the resistant bacteria because the infections caused by these organisms do not respond to routine antibiotics [1,3]. Eventually, newer drugs have to be produced to combat the drug resistant bacterial infections.
Earliest incidences of drug resistance were observed in 1948, when 65-85% of staphylococcus isolated were β-lactamase producers and were resistant to Penicillin G (Benzyl penicillin). In 2004, β-lactamase resistant penicillins were temporarily used, but soon nafcillin resistant Staphylococcus too has emerged leading to the use of vancomycins. But soon vancomycin resistant strains of Staphylococcus were isolated from the patients. Plasmid causing drug resistant genes occurs in many gram-negative bacteria of the normal gut flora. The abundant use of antibiotics suppresses the growth of normal bacteria and promotes the growth of klebsiella, enterobacter, proteus and pseudomonas [7,8].
Recently, drug resistance has evolved to a more advanced form as MDR. MDR is the resistance of microorganism to at least one agent in three or more antimicrobial classes [4].
These drug resistant bacteria like any other bacteria colonise the denture surface which left uncleaned, will mature over a period of time and dissipate free planktonic bacteria over distant sites which cause serious life threatening infections [3,9-11]. Hence, it is essential to study the drug resistant potential of some selective bacteria colonising the oral cavity as well as denture surfaces.
Scanning Electron Microscopy (SEM) has been used from early days for the purpose of examination and characterisation of bacteria found on medical devices [11,12]. SEM has the level of magnification and resolution which enables the observation of not only the shape of microorganisms, but also spatial organisation [11-14]. Hence, by using SEM, the surface morphological features of the bacteria along with bacterial membranes could be observed. This valuable data could be used as a guide for verification of effectiveness of antibiotics/antibacterial agents used against these drug resistant bacteria in future studies. This present study was conducted with a aim of isolation and detection of drug resistant bacteria in the complete denture patients including visualisation of drug resistant bacteria namely Staphylococcus aureus, Viridians streptococcus, Klebsiella pneumoniae and E.coli using SEM.
Materials and Methods
The study was a descriptive study and the design was laboratory invitro study design. Institutional ethics committee approval (Approval no.: IHEC/0142/2016) was obtained and study was carried out. Convenient sampling method was opted. Patients visiting the OPD of Rajahmuthiah Dental College and Hospital, Annamalai University, Chidamabaram, Tamil Nadu, India were selected for the study. The sample collection was done between June 2016 to December 2016. The study was carried out from June 2016 to December 2016.
Sample Size Calculation
The sample size was determined using Fisher’s (1998) [15] formula for sample size determination;
n=Z2PQ/D2 where, n=Desired sample size population.
Z=standard normal deviate set 1.96 for 95% confidence interval
P=isolation frequencies of the bacteria from the subjects from pilot study (0.11)
Q=1-P=0.89, D=Margin of error set as (0.08)
Hence n=1.962×0.11×0.89/(0.08)2=29.98 (rounded off to 30)
Hence sample size fixed at 30.
Inclusion Criteria: Healthy patients (i.e., no systemic diseases) and who were complete denture wearers at least for the past six months were included.
Exclusion criteria:
1) Patients with chronic systemic diseases and who were on medications.
2) Patients who had used local or systemic antibiotics within 30 days prior to sample collection.
3) Patients who had smoking habit.
Sample collection: An informed consent was obtained from the patients. A sterile swab (Hiculture transport swab, Himedia, Mumbai, India) was used to collect samples from patient’s oral cavity as well as from all over the complete denture’s surface. The patient’s denture surfaces namely occlusal surfaces, tissue surfaces as well as polished surfaces were swabbed. After swabbing the denture surfaces, the palatal, buccal and tonsillar surfaces of the intraoral cavity as well as the dorsal/lingual surfaces of the tongue were swabbed. The swab was subjected to microbiological investigation in the laboratory.
Microbiological Procedures
Selective media were obtained from Himedia laboratories, Mumbai, India, and they were prepared and sterilised using autoclave and poured in petriplates. The selective media used for Staphylococcus aureus, Viridians streptococcus, Klebisella pneumoniae, E.coli species were Mannitol salt agar, Mutans-Sanguis Agar, klebsiella selective agar and Eosin Methylene Blue (Himedia laboratories, Mumbai, India), respectively. The procedure involves inoculation of 10 μL of sample in sterile selective media plates by spread plating method and incubated at 37°C for 24 hours initially [16,17] and observation were made for changes until 48 hours.
Based on colony morphology, the colonies were selected and subcultured on sterile nutrient agar plate by quadrant streaking which was followed by gram staining of the pure isolate by using Gram stain Kit (Himedia Mumbai, India, Lot number: 0000187211). The stained slide was observed under 100X oil immersion objective using Bright field microscope (Apex industrial electronics, Haryana, India) to study the morphology. Based on similar characteristics in gram staining, the bacteria were selected and biochemical tests [17,18] were done to confirm the microorganism.
Steps Involved in Identification of Microbial Culture using 16S rRNA based Molecular Technique
Genomic DNA was isolated and quantity was measured using Nanodrop Spectrophotometer 2000/2000c (Thermo-fisher scientific, USA).
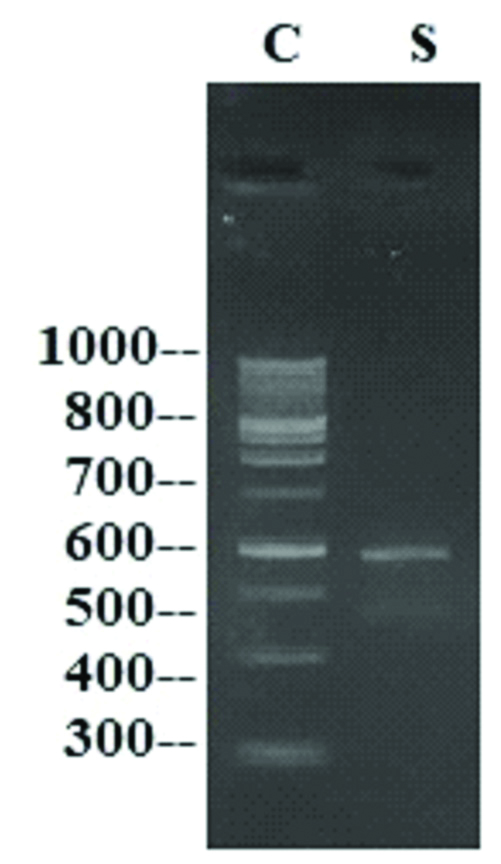
16S rRNA gene was amplified by 16S rRNA forward and reverse primers. A single discrete PCR amplicon band was observed when resolved on Agarose gel [Table/Fig-1] [19].
Representative image of PCR amplicon on Agarose gel (C represents 100 base pairs DNA ladder, S represents sample lane).
Forward and reverse DNA sequencing reaction of PCR amplicon was carried out with forward primer and reverse primers using BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyser (Applied Biosystems, Thermo Fischer Scientific).
Consensus sequence of 16S rRNA gene was generated from forward and reverse sequence.
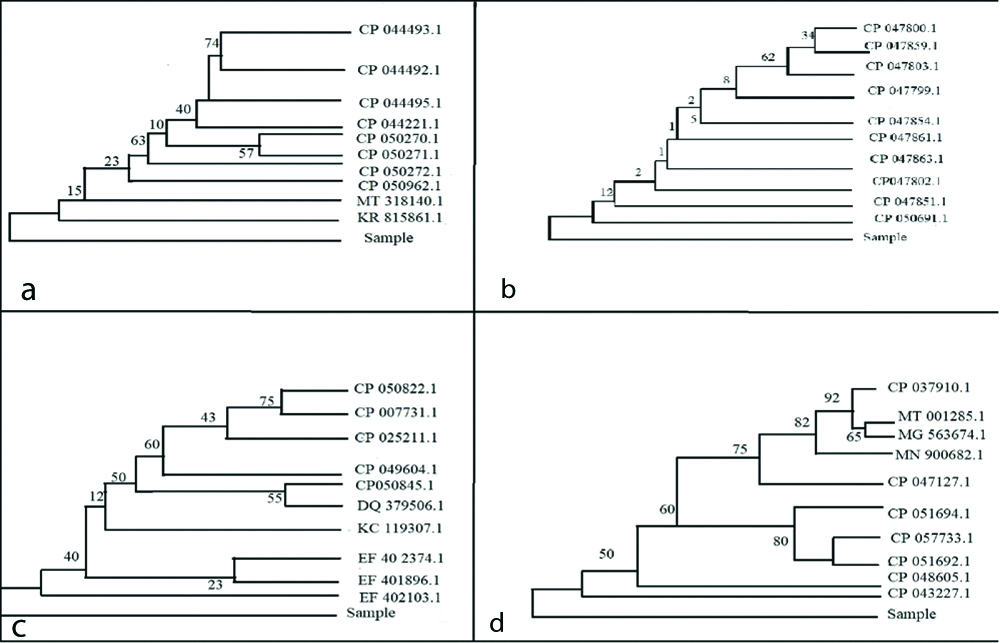
16S rRNA gene sequence was used to carry out BLAST with the database of NCBI genbank database [20,21]. Based on maximum identity score, first ten sequences were selected and aligned using multiple alignment software program Clustal W (https://embnet.vital-it.ch/software/ClustalW.html). Distance matrix was generated and the phylogenetic tree [Table/Fig-2] was constructed by using MEGA7 [22].
Phylogenetic trees of viridans streptococci species (a), Staphylococcus aureus (b), Klebsiella pneumoniae (c), E.coli (d).
From the total 30 samples taken for the study, only 20 isolates of each of the selected bacteria were ascertained by the 16S rRNA and biochemical tests. These isolates were in turn subjected to antibiotic sensitivity tests.
Antibiotic Sensitivity Test
After microbiological tests and 16S rRNA sequencing the pure isolates were subjected to antibiotic sensitivity by agar disc diffusion method on Muller Hinton Agar (MHA) medium. The inoculums were spread on the MH Agar with sterile swab moistened with the bacterial suspension and antibiotic discs were placed [16-18]. These plates were incubated for 24 hours at 37°C. Then the microbial growth was determined by measuring the diameter of zone of inhibition by using antibiotic zone scale (Himedia, India).
The zone of inhibition was measured from one edge of the zone to the other edge across the centre of the antibiotic disc. Antibiotic disks used for antibiotic sensitivity for Staphylococcus aureus and Viridans streptococcus species included were: Amoxicillin (10 μg) (AM), Amoxicillin Clavulanic Acid (10 μg) (AMC/CLAV), Cefotaxime (30 μg) (CEF), Gentamycin (10 μg) (GEN), Methicilin (METH), Doxicillin (30 μg) (DOX) and Clindamycin (2 μg) (CLIN). Methicillin resistance was found by means of oxacillin (1 μg) [23].
Antibiotic disks used for antibiotic sensitivity for Klebsiella pneumoniae and E. coli included were AM (10 μg), AMC/CLAV (10 μg), CEF (30 μg), GEN (10 μg), Ofloxacin (OFL) (5 μg), DOX (30 μg), Chlaramphenicol (CHLO) (30 μg).
Criteria for Drug Resistance
The zone of inhibition was measured using ruler across the centre of antibiotic disks. Isolates were classified as Sensitive, Intermediate and Resistant based on CLSI guidelines [5] as represented in [Table/Fig-3].
Zone of inhibition for various antibiotics used in the study.
| Antibiotics | Viridans streptococci | Staphylococcus aureus | Klebsiella pneumoniae | E.coli |
|---|
| Sen | Int | Res | Sen | Int | Res | Sen | Int | Res | Sen | Int | Res |
|---|
| Amoxyclav (10 μg) | 18 | 14-17 | 13 | 20 | | 19 | 18 | 14-17 | 13 | 18 | 14-17 | 13 |
| Ampicillin (10 μg) | 21 | 15-20 | 15 | 29 | - | 28 | 17 | 14-16 | 13 | 17 | 14-16 | 13 |
| Cefotaxime (30 μg) | 28 | 26-27 | 25 | 23 | 15-22 | 14 | 26 | 23-25 | 22 | 26 | 23-25 | 22 |
| Chloramphenicol (30 μg) | 21 | 18-20 | 17 | 18 | 13-17 | 12 | 18 | 13-17 | 12 | 18 | 13-17 | 12 |
| Clindamycin (2 μg) | 19 | 16-18 | 15 | 21 | 15-20 | 14 | 17 | 15-16 | 14 | 17 | 15-16 | 14 |
| Doxycyclin (30 μg) | 12 | 13-15 | 16 | 16 | 13-15 | 12 | 14 | 11-13 | 10 | 14 | 11-13 | 10 |
| Gentamycin (10 μg) | 13 | 14-22 | 23 | 15 | 13-14 | 12 | 15 | 13-14 | 12 | 15 | 13-14 | 12 |
| Methicilin (oxacilin 1 μg) | | | | 13 | 11-12 | 10 | | | | | | |
| Ofloxacin (5 μg) | 16 | 13-15 | 12 | 18 | 15-17 | 14 | 16 | 13-15 | 12 | 16 | 13-15 | 12 |
Sen: Sensitive; Int: Intermediate; Res: Resistant measured in millimeters
Criteria for Multidrug Resistance (MDR)
Isolates were categorised as multidrug resistant if resistance is shown to atleast one antibiotic agent in three or more antibiotic group [4].
Fabrication of Acrylic Strips
Transparent denture base acrylic resin strips (DPI, Mumbai, India) were fabricated by means of wax patterns [24] with the dimensions as length 50 mm, width 10 mm, and thickness 2 mm. The wax samples were prepared using custom-made metal moulds. The wax strips (Brulon international, Valsad, India) were then invested in dental flasks with type III gypsum product (Asian chemicals, Rajkot, India), i.e., dental stone, and dewaxing was done. The heat cure acrylic resin of polymer: monomer ratio of 3:1 by volume was prepared and polymerised with by means of conventional flasking technique following long curing cycle. Then the acrylic strips were reduced to a length 8 mm, breadth 8 mm and thickness 2 mm for visualising the drug resistant bacteria via SEM. This was done to accommodate the holder dimensions in scanning electron microscope.
Scanning Electron Microscope (SEM) Study
The biofilms were grown overnight on a sterilised acrylic strip. The samples were then rinsed in 0.1 M buffered sodium cacodylate (Merck KGaA, Darmstadt, Germany) and dehydrated by serial transfers in various concentrations of ethyl alcohol (Krishna Pharma, Hyderabad, India) for 30 minutes each [25]. After 24 hours at room temperature, the specimens were mounted on metal stubs, coated with a layer of gold under vacuum by sputter coating machine (JEOL, JPC 1600, JEOL companies, Japan). After the gold coating the specimens were visualised by scanning electron microscope (JEOL, JEOL Ltd., Tokyo, Japan) operated at 15 Kilovolts.
Results
The range of the age of sample population selected for the study was between 45-70 years. Male subjects were 18 and females were 12 in number.
The isolated bacteria were subjected to agarose gel and individual PCR amplicons were obtained whose base pairs closely resemble base pairs of respective bacteria. Representative image of PCR amplicons given in [Table/Fig-1] where PCR amplicons of 600 base pairs are clearly seen.
The results of the 16S rRNA sequencing suggested that the isolated bacterial samples are closely related to evolutionary taxa of their respective bacterial strains [Table/Fig-2].
The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches [Table/Fig-2]. For Viridans streptococcus species 57 associated taxa were clustered around strains with accession numbers CP 050270.1 and CP 050271.1 [Table/Fig-2a]. With regard to Staphylococcus aureus a maximum clustering of associated taxa (n=62) was for Strain (Accession no. CP 047803.1) [Table/Fig-2b]. The phylogenetic tree of Klebsiella pneumoniae reveals that there were 75 associated taxa clustered around strains CP 050822.1 and CP 007731.1 [Table/Fig-2c]. E.coli phylogenetic tree maximum clustering of taxa numbering 92 around strain CP 037910.1 [Table/Fig-2d].
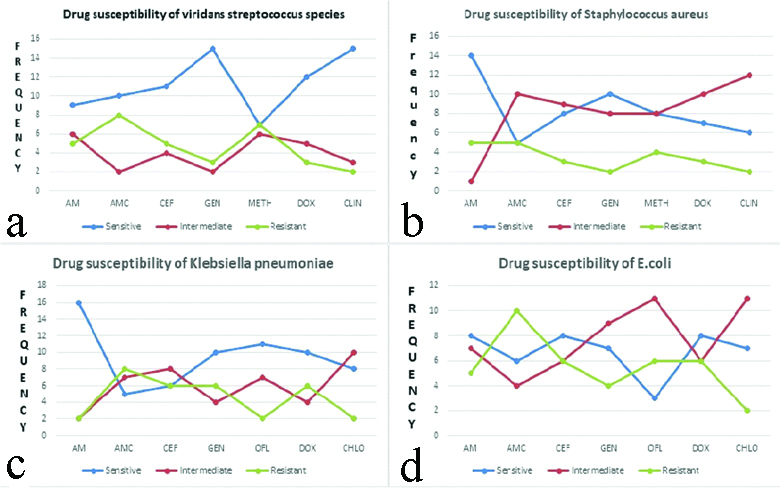
Zone of inhibition for various antibiotics used in the study is shown in [Table/Fig-3]. Highest percentages of Viridans streptococcus isolates which exhibited drug resistance was for AMC/CLAV and METH which accounted for 40% and 35% of the isolates, respectively. Drug resistance was found to be least for DOX and CLIN which accounted for 15% and 10%, respectively. About 75% of the Viridans streptococcus isolates were susceptible to both GEN and CLIN [Table/Fig-4,5a-d].
Viridans streptococcus species, Staphylococcus aureus, Klebsiella pneumoniae and E.coli isolates susceptibility to antibiotics.
| Bacteria | Number of Isolates Sensitive, Intermediate and Resistant Isolates to Antibiotics |
|---|
| AM | AMC | CEF | GEN | METH | DOX | CLIN#/CHLO$ |
|---|
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R |
|---|
| Viridans streptococcus speciesN (%) | 9(45) | 6(30) | 5(25) | 10(50) | 2(10) | 8(40) | 11(55) | 4(20) | 5(25) | 15(75) | 2(10) | 3(15) | 7(35) | 6(30) | 7(35) | 12(60) | 5(25) | 3(15) | 15(75) | 3(15) | 2(10) |
| Staphylococcus aureusN (%) | 14(70) | 1(5) | 5(25) | 5(25) | 10(50) | 5(25) | 8(40) | 9(45) | 3(15) | 10(50) | 8(40) | 2(10) | 8(40) | 8(40) | 4(20) | 7(35) | 10(50) | 3(15) | 6(30) | 12(60) | 2(10) |
| Klebsiella pneumoniaeN (%) | 16(80) | 2(10) | 2(10) | 5(25) | 7(35) | 8(40) | 6(30) | 8(40) | 6(30) | 10(50) | 4(20) | 6(30) | 11(44) | 7(35) | 2(10) | 10(50) | 4(20) | 6(30) | 8(40) | 10(50) | 2(10) |
| E.coliN (%) | 8(40) | 7(35) | 5(25) | 6(30) | 4(20) | 10(50) | 8(40) | 6(30) | 6(30) | 7(35) | 9(45) | 4(20) | 3(15) | 11(55) | 6(30) | 8(40) | 6(30) | 6(30) | 7(35) | 11(55) | 2(10) |
(AM) Amoxicillin, (AMC) Amoxicillin clavulinic acid, (CEF) Cefotaxime, (GEN) Gentamycin, (METH) Methicilin, (DOX) Doxicillin, (CLIN) Clindamycin (#for viridans streptoccus spp. and S.aureus, (CHLO) Chloramphenicol ($for K.pneumoniae and E.coli)
a,b) Antibiotics disc in the media before incubation for Viridans streptococcus species. c,d) Antibiotics disc in the media after incubation for Viridans streptococcus species (Zone of inhibition indicated by white arrows).
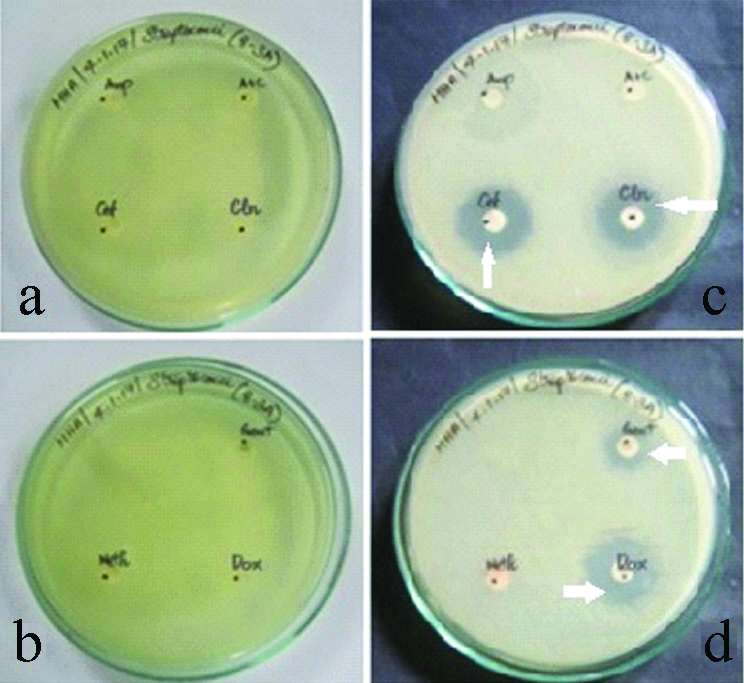
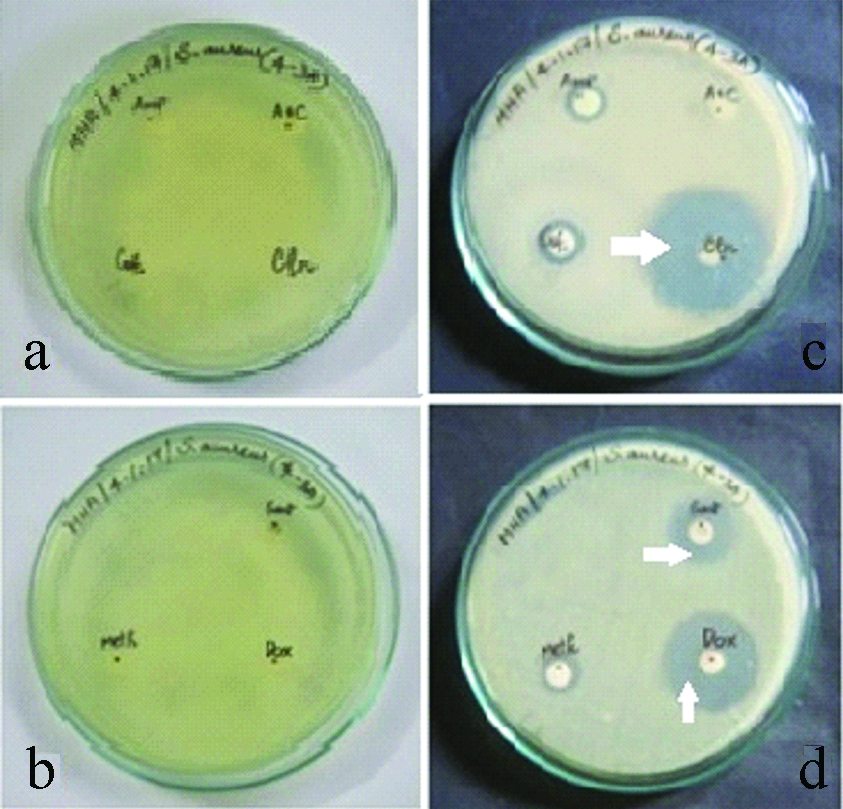
With regard to staphylococcus aureus 25% of the isolates were resistant to AM, AMC/CLAV, whereas least resistance was shown to GEN and CLIN which accounted for 10% isolates. The drug for which staphylococcus aureus isolates were most susceptible was AM which accounted for 70% of the S.aureus isolates [Table/Fig-4,6a-d].
a,b) Antibiotics disc in the media before incubation for Staphylococcus aureus. c,d) Antibiotics disc in the media after incubation for Staphylococcus aureus (Zone of inhibition indicated by white arrows).
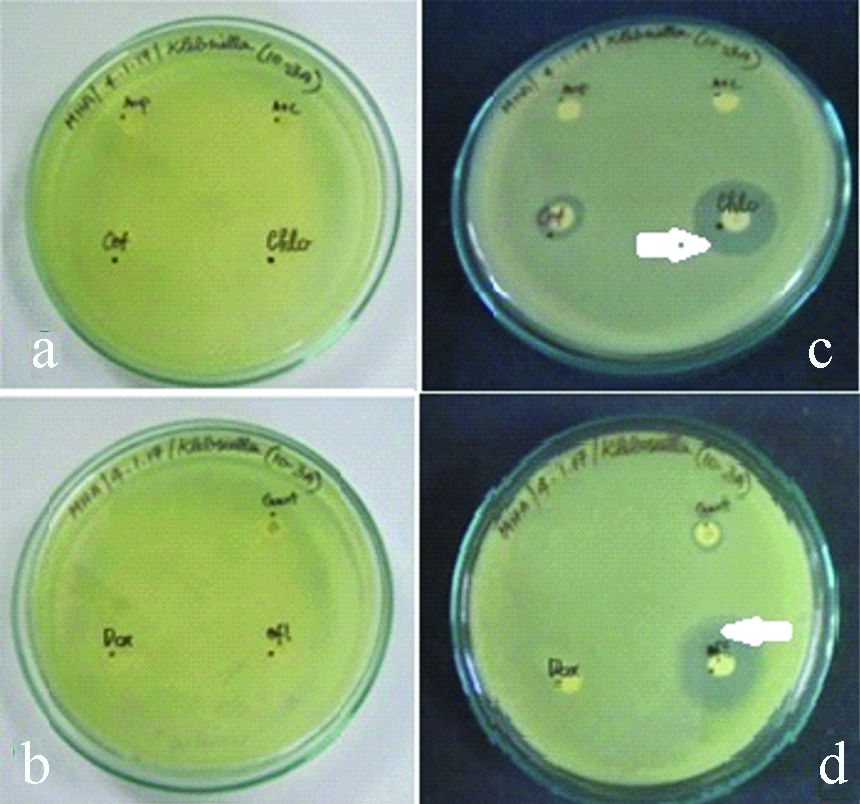
Drug resistance was found to be highest for AMC/CLAV with regard to 40% of Klebsiella pneumoniae isolates. About 30% of the isolates also exhibited drug resistance to CEF, GEN and DOX. Least drug resistance was found for METH and CHLO (10% K.pneumoniae isolates). Drug susceptibility was found to be highest for AM in about 80% of the K.pneumoniae isolates [Table/Fig-4,7a-d].
a,b) Antibiotics disc in the media before incubation for Klebsiella pneumoniae. c,d) Antibiotics disc in the media after incubation for Klebsiella pneumoniae. (Zone of inhibition indicated by white arrows).
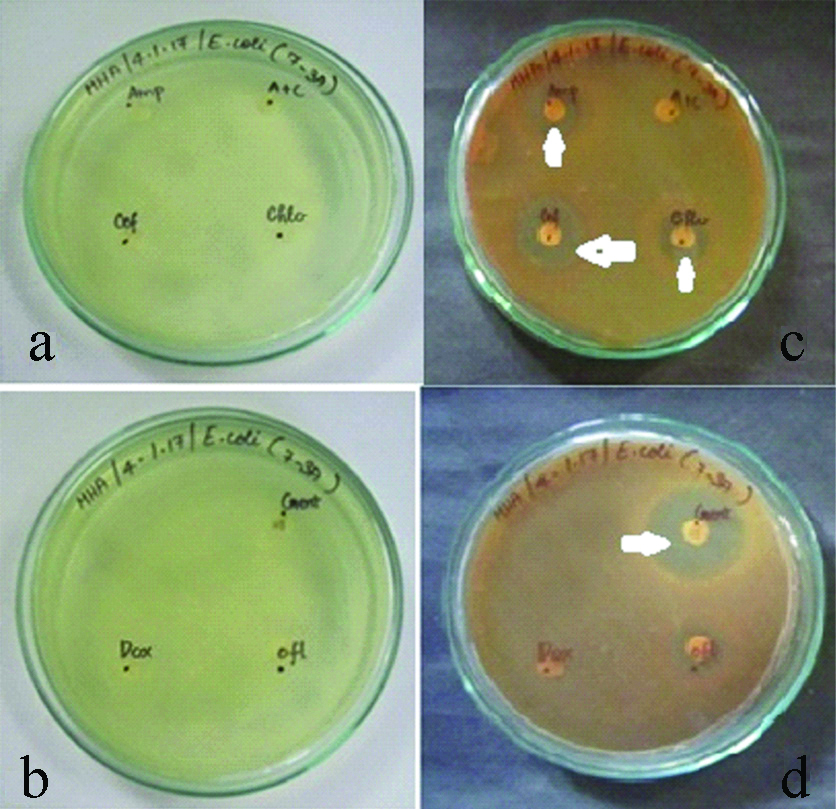
Highest percentages of E.coli isolate which showed drug resistance was for AMC which accounted for 50%. Drug resistance to CEF, METH and DOX was reveled by 30% of E.coli isolates. CLIN was found to be drug which is resistant in only 10% of the isolates. About 40% isolates were susceptible to AM, CEF and DOX [Table/Fig-4,8a-d]. SEM image of Viridians streptococcus, Staphylococcus aureus, Klebsiella pneumoniae and E. coli is represented in [Table/Fig-9a-d]. Drug susceptibility of Viridans streptococci spp., Staphylococcus aureus, Klebsiella pneumoniae, E.coli is shown in [Table/Fig-10a-d].
a,b) Antibiotics disc in the media before incubation for E.coli. c,d) Antibiotics disc in the media after incubation for E.coli. (Zone of inhibition indicated by white arrows).
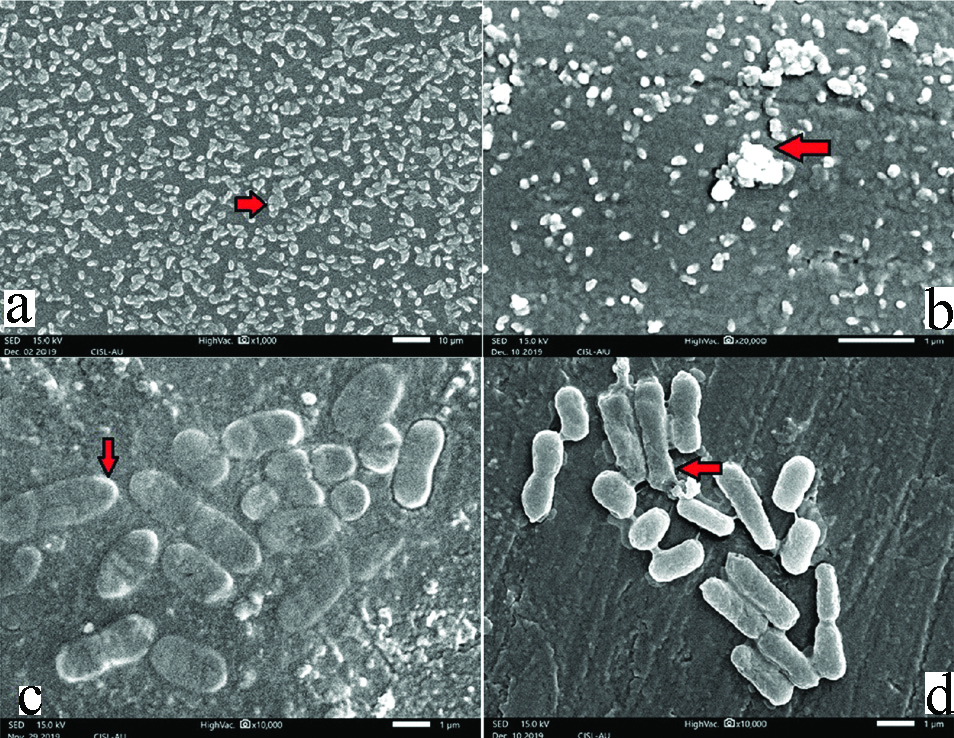
a) SEM image of Viridians streptococcus (red arrow indicates increased cell density). b) SEM image of Staphylococcus aureus (red arrow indicates increased cell density). c) SEM image of Klebsiella pneumonia (red arrow indicates thick capsule). d) SEM image of E. coli (red arrow indicates Extracellular Polysaccharide Substance (EPS).
Drug susceptibility of Viridans streptococci spp. (a), Staphylococcus aureus (b), Klebsiella pneumoniae (c), E.coli (d).
Discussion
In the present study drug resistance was found in each of the selected bacteriae for the antibiotics used in the study. The number of isolates of individual bacteria which proved to be drug resistant varied for different antibiotics.
In the present study, among the isolates of Viridans streptococcus species, 40% isolates showed resistance to AMC followed by 35% isolates to METH (oxacillin). This finding is much similar to a study conducted by Sweeney LC et al., wherein they found that high-level penicillin resistance was shown in 8% of S. salivarius strains, 20% of S. mitis strains and 35% of S. oralis strains [26].
With regard to Staphylococcus aureus 25% isolates showed resistance to both AM and AMC, whereas only 20% isolates showed resistance to METH (oxacillin). A study conducted by Yamashita K et al., have documented a higher percentage of staphylococcus aureus which were resistant to METH (oxacillin) which was 69.2% in oral cancer patients as against the present study which was only 25% for healthy edentulous patients [27].
The SEM images obtained in the present study are very much similar to study done by Gomes LC and Mergulhäo FJ who have attributed increased cell density [14] and/or presence of EPS to the presence of drug resistance [26]. The spatial arrangement of a higher number of cells could create concentration gradients (of nutrients, antibiotic, and oxygen) within the biofilm structure that could contribute to drug resistance [28,29].
About 66.7% of the Klebsiella pneumoniae isolates from complete denture patients showed resistance to AM and AMC/CLAV according to Gaetti-Jardim EC et al., whereas in the present study 40% isolates of Klebsiella pneumoniae showed resistance to AMC followed by GEN, CEF for which 30% of the isolates have shown resistance [30,31]. The presence of EPS around the bacterial cells could be the reason behind the drug resistance of K. pneumoniae as EPS acts as protective layer preventing the penetration of antibiotics. This was observed in a SEM study conducted by Bandeira M et al., [32]. Another reason for increased drug resistance is the presence of capsule which is clearly evident in our present study via SEM image. Gaetti-Jardim EC et al., [29,30] have also found that 33.3% isolates of E.coli have resistance to Doxicillin which is very much similar to the present study. It was also found in the present study that in 30% of the E.coli isolates exhibited resistance to CEF and OFL. E.coli biofilms which were drug resistant were observed through SEM studies by Gomes LC and Mergulhäo FJ, which was much similar to this study wherein thick EPS binding the individual’s cells were observed in the SEM images [14].
Gaetti-Jardim EC et al., also have found that none of the isolates of Klebsiella pneumoniae and E.coli were resistant to gentamycin [29,30,32]. Hence, Gentamycin could be a drug of choice for Enterobacteriaceae group of bacteria which are resistant to other antibiotic groups used in the treatment of infections caused by Enterobacteriaceae namely urinary tract infections, respiratory and gastrointestinal infections.
Limitation(s)
Sample size was restricted to 30 as wide array of microbiological, sequencing and antibiotic sensitivity testing was done. Study was restricted to healthy patients only. The study could be extended to immunocompromised patients namely patients with diabetes mellitus, HIV, patients who are undergoing chemotherapy, on dialysis, transplants etc.
Conclusion(s)
From the study it has been observed that drug resistant gram-positive bacteria (Viridans streptococcus spp.) and gram-negative bacteria, (Klebsiella pneumoniae, E.coli) have been detected from the complete denture patients.
By means of the high-resolution imaging of biofilm surfaces, it could be concluded that SEM is a valuable tool to study the drug resistant biofilm. Antibiotic sensitivity tests, 16S rRNA sequencing and SEM studies are valuable tools to study dental biofilms especially those biofilms which are specifically drug resistant. Cell density, EPSs and capsule could be important factors for providing drug resistance. Inclusion of a greater number of samples from both healthy and non healthy population could be done in future studies to know the prevalence and categorisation of drug susceptibility.
(AM) Amoxicillin, (AMC) Amoxicillin clavulinic acid, (CEF) Cefotaxime, (GEN) Gentamycin, (METH) Methicilin, (DOX) Doxicillin, (CLIN) Clindamycin (#for viridans streptoccus spp. and S.aureus, (CHLO) Chloramphenicol ($for K.pneumoniae and E.coli)