Hence, analysis of WHONET surveillance data was undertaken to know the aetiological profile of uropathogens and their antimicrobial susceptibility pattern to formulate an empirical treatment policy in a tertiary care hospital.
Materials and Methods
The present study was retrospective and cross-sectional. It was conducted in the Government Medical College Hospital Nagpur, a tertiary care hospital in Central India. The data generated during the period July 2018 to June 2019 was subsequently analysed. All urine specimens received in the laboratory for culture were processed and isolated uropathogens were analysed for their antimicrobial susceptibility. Analysis of Multi-Drug Resistant (MDR) uropathogens was carried out using WHONET software.
The urine specimens were processed by standard microbiological techniques and the uropathogens were identified by conventional methods [5]. Antimicrobial susceptibility testing was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines using the Kirby-Bauer disc diffusion technique [6]. An inclusion criterion was uropathogens with significant growth of one pathogen and only the first isolate of a given species encountered in case there were repeat samples from the same patient [7]. The software has the option to choose the first isolate of a patient. Samples with insignificant growth were excluded.
The data entry and analysis was done using the software WHONET 5.6 version [8]. The software was customised with respect to patient’s location in the hospital and the antimicrobials used in the susceptibility testing along with their strengths. Quality control of culture media and antibiotic discs was done by standard protocols and is a regular feature of the laboratory. Reproducibility of the sensitivity test results were checked regularly at periodic intervals. CLSI 2013 recommended 3×5 day quality control plan [9] which was followed before implementing to a weekly schedule which is the ongoing internal quality control policy of the department.
Statistical Analysis
Categorical variables were expressed in terms of frequencies and percentages. Chi-square test was used to compare percentages. The p-value <0.05 was considered as statistically significant. Statistical analysis was performed by using Open Epi version 3.
Results
A total of 1152 (13.4%) non-duplicate uropathogens were isolated from 8573 patients of UTI. In culture positive patients, 527 (46%) were male, 618 (53%) female and 7 (1%) were neonates who were identified by the name of their mother; hence their sex was not included in data entry. There were 1081 (94%) adults, 64 (5%) paediatric patients and 7 (1%) neonates (data of age of the patients was not available). Outpatients were 149 (13%) and the remaining 1003 (87%) hospitalised (indoor) patients included 110 (10%) from ICU and 893 (77%) from non-ICU wards. Department-wise distribution of uropathogens was 36% from General Medicine, 7% from Paediatrics, 16% from Nephrology, 7% from General Surgery, 7% from Urology, 16% from Obstetrics and Gynaecology and collectively 11% from other departments which included Cardiovascular Thoracic Surgery, Endocrinology, Gastroenterology, Infectious Diseases, Burn, Neurology, Ophthalmology, Orthopaedics, Paediatric Surgery, Radiotherapy, Respiratory Medicine, Skin and Venereal Disease, Trauma Care Centre.
Different uropathogens isolated and their distribution in major departments is shown in [Table/Fig-1]. It shows that maximum isolates were from Medicine (36%) followed by Nephrology and Obstetrics and Gynaecology (16% each). Enterobacteriaceae was the commonest causative agent in all the departments. Chi-square test has been applied to compare major microbial groups in different departments.
Aetiological profile of uropathogens in major clinical departments.
| Organisms | No. of isolates in different departments (%) | Chi square test | Degree of freedom | p- value |
|---|
| Medicine | Nephrology | Surgery | Urology | Obstetrics and gynaecology | Paediatrics | Othersφ | Total (%) |
|---|
| GPC@ | 56 (14) | 18 (10) | 9 (12) | 9 (11) | 28 (15) | 12 (15) | 20 (15) | 152 (13) | 3.08 | 5 | 0.68 |
| S. aureus | 6 (2) | 3 (2) | 1 (1) | 3 (4) | 5 (3) | 4 (5) | 3 (2) | 25 (2) |
| E. faecalis | 50 (12) | 15 (8) | 8 (11) | 6 (7) | 23 (12) | 8 (10) | 17 (13) | 127 (11) |
| EBC# | 299 (73) | 142 (78) | 59 (78) | 60 (73) | 141 (76) | 58 (73) | 101 (76) | 860 (75) | 2.30 | 5 | 0.81 |
| E. coli | 222 (54) | 108 (59) | 38 (50) | 44 (54) | 108 (58) | 44 (55) | 75 (56) | 639 (55) |
| K. pneumoniae | 72 (18) | 31 (17) | 21 (28) | 12 (15) | 29 (16) | 13 (16) | 23 (17) | 201 (17) |
| Pr. mirabilis | | | | 1 (1) | | | 2 (2) | 3* |
| Citro. koseri | 5 (1) | 3 (2) | | 3 (4) | 4 (2) | 1 (1) | 1 (1) | 17 (1) |
| NFR$ | 45 (11) | 21 (12) | 7 (9) | 12 (15) | 18 (10) | 9 (11) | 9 (7) | 121 (10) | 1.76 | 5 | 0.88 |
| P. aeruginosa | 26 (6) | 16 (9) | 4 (5) | 11 (14) | 7 (4) | 5 (6) | 4 (3) | 73 (6) |
| Acineto baumannii | 19 (5) | 5 (3) | 3 (4) | 1 (1) | 11 (6) | 4 (5) | 5 (4) | 48 (4) |
| Candida | 11 (2) | 2 (1) | 1 (1) | 1 (1) | | 1 (1) | 3 (2) | 19 (2) | 1.19 | 4 | 0.87 |
| Total | 411 (36) | 183 (16) | 76 (7) | 82 (7) | 187 (16) | 80 (7) | 133 (11) | 1152 (100) | | | |
1. Abbreviations and classes of uropathogens is as per the WHONET version 5.6.
2. @Gram positive cocci, #Enterobacteriaceae, $Non-fermenting gram negative bacilli, *Only 3 isolates of Pr mirabilis accounted for <1%, φOthers include Cardiovascular Thoracic Surgery, Endocrinology, Gastroenterology, Infectious Diseases, Burn, Neurology, Ophthalmology, Orthopaedics, Paediatric Surgery, Radiotherapy, Respiratory Medicine, Skin and Venereal Disease, Trauma Care Centre departments.
Percentage values are rounded off and not given in decimals. Little adjustments are made to get total as 100%.
4. By statistical test, no. of isolates of microbial groups in different departments are compared.
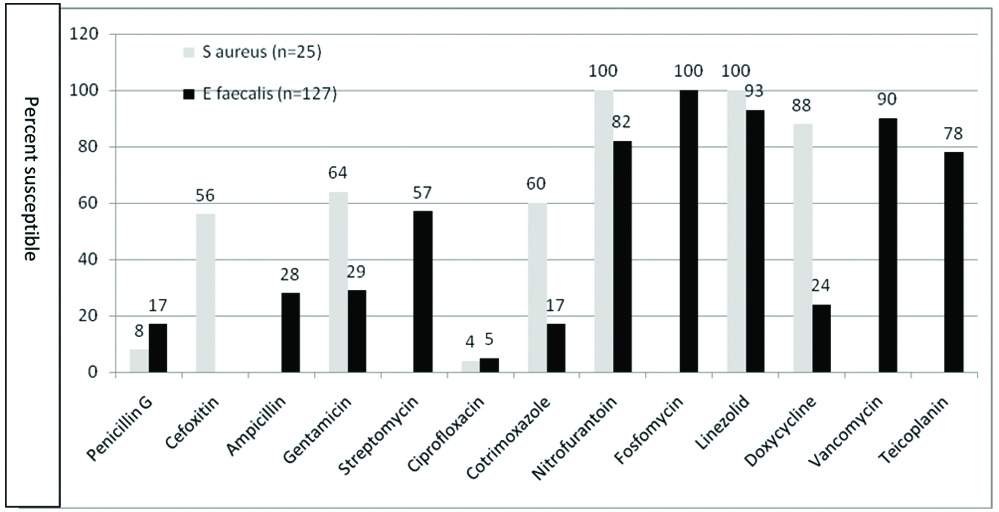
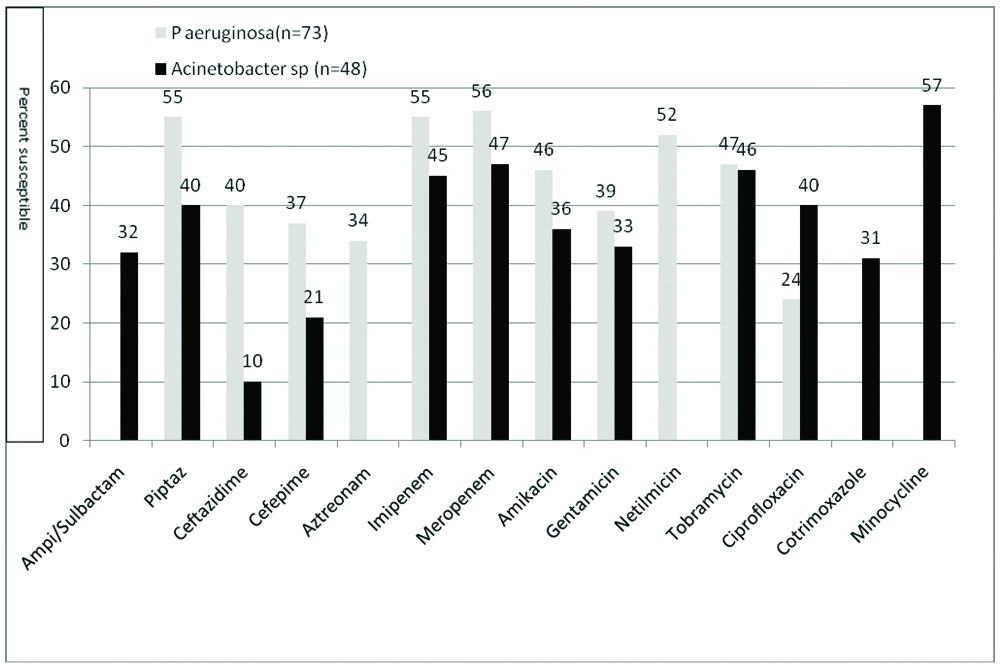
A result of antimicrobial susceptibility of Gram positive cocci (S. aureus and E. faecalis) is shown in [Table/Fig-2]. Both S. aureus and E. faecalis showed high sensitivity for nitrofurantoin and linezolid and low sensitivity for ciprofloxacin and penicillin G. [Table/Fig-3] shows the antimicrobial susceptibility of Enterobacteriaceae. As compared to K pneumoniae, E coli showed higher susceptibility for most of the antibiotics. Chi-square test has been applied to compare susceptibility of E coli and K pneumoniae for different antibiotics. All the β-lactam antibiotics showed high resistance except piperacillin-tazobactam and carbapenem. A result of antimicrobial susceptibility of Nonfermenters (P. aeruginosa and A. baumannii) is shown in [Table/Fig-4]. They showed low susceptibility to most of the antibiotics tested.
Antimicrobial susceptibility of S. aureus and E. faecalis isolated from UTI (%).
Antimicrobial susceptibility of Enterobacteriaceae isolates from UTI.
| Name of antibiotic | No of susceptible strains (%) | Chi square test | Degree of freedom | p-value |
|---|
| E coli (n=639) | K pneumoniae (n=201) | Enterobacteriaceae (n=860) |
|---|
| Ampicillin | 47 (07) | 0 (00) | 50 (06) | 15.66 | 1 | <0.001 |
| Amoxiclav | 76 (12) | 14 (07) | 93 (11) | 3.88 | 1 | 0.048 |
| Piperacillin tazobactam | 358 (56) | 68 (34) | 433 (50) | 30.13 | 1 | <0.001 |
| Cefazolin | 79 (12) | 12 (06) | 94 (11) | 6.46 | 1 | 0.011 |
| Cefuroxime | 79 (12) | 12 (06) | 94 (11) | 6.46 | 1 | 0.011 |
| Ceftazidime | 122 (19) | 16 (08) | 141 (16) | 13.80 | 1 | <0.001 |
| Cefotaxime | 90 (14) | 16 (08) | 109 (13) | 5.20 | 1 | 0.022 |
| Cefepime | 128 (20) | 18 (09) | 152 (18) | 13.06 | 1 | <0.001 |
| Cefoxitin | 135 (21) | 22 (11) | 159 (18) | 10.42 | 1 | 0.0012 |
| Imipenem | 358 (56) | 68 (34) | 433 (50) | 30.13 | 1 | <0.001 |
| Meropenem | 435 (68) | 75 (37) | 520 (60) | 60.66 | 1 | <0.001 |
| Amikacin | 403 (63) | 99 (49) | 511 (59) | 12.13 | 1 | 0.0004 |
| Gentamicin | 294 (46) | 83 (41) | 388 (45) | 1.37 | 1 | 0.24 |
| Tobramycin | 352 (55) | 85 (42) | 453 (53) | 10.03 | 1 | 0.0015 |
| Ciprofloxacin | 96 (15) | 18 (09) | 118 (14) | 4.8 | 1 | 0.028 |
| Levofloxacin | 116 (18) | 43 (21) | 177 (21) | 1.04 | 1 | 0.306 |
| Norfloxacin | 84 (13) | 18 (09) | 106 (12) | 2.51 | 1 | 0.112 |
| Cotrimoxazole | 192 (30) | 64 (32) | 265 (31) | 0.23 | 1 | 0.630 |
| Fosfomycin | 627 (98) | Not tested | 627 (98)* | | | |
| Nitrofurantoin | 544 (85) | 115 (57) | 664 (77) | 70.50 | 1 | <0.001 |
1. Percentage susceptibility of Proteus mirabilis and Citrobacter koseri has not been given, as the no. of isolates are very few. 2. *Fosfomycin tested for E coli only, 627 strains of E coli are susceptible from its total 639 isolates. 3. By statistical test, for different antibiotics susceptibility of E coli and K pneumoniae is compared.
Antimicrobial susceptibility of P. aeruginosa and A. baumannii isolates from UTI.
Discussion
Global surveillance of antimicrobial resistance is one of the goals of WHO. To support the goal, WHO has developed an information system, WHONET. This software facilitates customisation of its components, analysis of data and networking locally, nationally and globally. WHONET helps in routine microbiology laboratory data management and also provides information about antimicrobial susceptibility patterns at different places over different time scales. It helps the administrator to assess and plan the antimicrobial policy of the hospital [10,11]. The software has many applications. The data for each individual organism, ward, area and for a wide range of antibiotics could be analysed. The month wise data could be analysed and the trends of each month of the year, along with the area and ward with infections could be compared. Identification of epidemic or nosocomial strains in the ward could be traced. These observations could help the Health Administrators of the Hospital, to take preventive actions depending on the season, ward and unit. Special areas could be easily identified and could help the infection control officer to take necessary action urgently when required. The data can be easily presented during hospital infection control meetings to directly advise the individual clinician to take corrective actions. It helps in formulation of empirical antimicrobial treatment policy for the institution [12,13].
Urinary tract is the most common organ system to experience bacterial infections. UTIs are challenging, not only because of the large number of infections that occur each year, but also due to the drug resistance in uropathogens. Class Enterobacteriaceae was the causative agent in 75% cases, E. coli accounting for more than two third of the cases. Classes Gram positive cocci and Non-fermenters were isolated in 13% and 11% cases, respectively [Table/Fig-1]. E. coli and Enterococci are the commensals of gastrointestinal tract hence they easily invade the urinary tract and therefore a common cause of UTI [14].
In the present study, the commonest uropathogen as shown in [Table/Fig-1] was E. coli (55%) followed by K. pneumoniae (17%) and E. faecalis (11%). Present study findings were in concordance with earlier reports [15-17]. It has been established that enterococcal colonisation of foley’s catheter among hospitalised patients plays a role in increasing the risk of infection due to enterococci [18], though in the present study E. faecalis was isolated in both catheterised and non-catheterised patients. Pathogens like S. aureus, P. aeruginosa and A. baumannii are known to circulate in hospital environment and are common cause of hospital acquired UTI [19]. Although 87% of the patients in the present study were indoor patients, the isolation of these pathogens was only 2%, 6% and 4%, respectively. This may be attributed to effective infection control practices in our hospital. Gupta K et al., reported coagulase negative staphylococci (S. saprophyticus) as one of the cause of community acquired UTI [17]. In the present study, 13% patients were from various OPDs. These patients were attending speciality clinics like antenatal, nephrology and urology OPDs on a regular basis. Patients attending nephrology OPD were on dialysis while urology OPD patients were post-procedural patients. These might not truly represent patients with community acquired UTI. This may be the plausible reason for not isolating coagulase negative staphylococci in this study. The antimicrobial resistance in uropathogens is increasing in both outpatients as well as hospitalised patients [14]. Understanding the impact of drug resistance is of critical importance as the changing rate of antibiotic resistance has a large impact on the therapy of UTI.
Antimicrobial susceptibility of Gram positive cocci in urinary isolates is shown in [Table/Fig-2]. Both S. aureus and E. faecalis showed high sensitivity to linezolid (100% and 93%, respectively) and to nitrofurantoin (100% and 82%, respectively). Shevade S and Agrawal G in their study of uropathogens reported high sensitivity of staphylococci and enterococci to linezolid (100% to both) and to nitrofurantoin (86% and 77%, respectively) [14]. Wandre A and Agrawal G in their study of clinical isolates of S aureus observed 100% sensitivity to both linezolid and nitrofurantoin [20]. Nanoty VV et al., reported 100% sensitivity of S. aureus to linezolid [21]. Rahangdale V et al., observed high sensitivity of enterococci to linezolid and nitrofurantoin (100% and 87.24%, respectively) [22]. While, nitrofurantoin is a good choice for empirical treatment for gram positive cocci in urine, linezolid must be kept as a reserve drug for serious MDR staphylococcal infections and also for MDR and Extensively Drug Resistant (XDR) tuberculosis [23]. Vancomycin and teicoplanin are currently in use for the treatment of infections caused by invasive β-lactam resistant gram positive cocci. The use of vancomycin and teicoplanin in UTI must therefore be discouraged. Further in staphylococcal isolates, sensitivity testing of glycopeptides requires Minimum Inhibitory Concentration (MIC) determination, which was not undertaken for urinary isolates, hence not reported in the present study. E. faecalis showed good sensitivity to fosfomycin (100%), vancomycin (90%) and teicoplanin (78%). In UTI, fosfomycin should not be the choice for empirical treatment, as fosfomycin testing is recommended only for urinary E coli and urinary E. faecalis [6].
In the class Enterobacteriaceae, around 98% isolates were either E. coli or K. pneumoniae. E. coli showed significantly higher susceptibility to most of the antimicrobials as compared to K. pneumoniae (p-value for all antimicrobial tested was <0.05 except levofloxacin, norfloxacin and Cotrimoxazole as shown in [Table/Fig-3]. Shewade S and Agrawal G also observed higher susceptibility in E coli as compared to K. pneumoniae [14]. Though, E. coli is 100% sensitive to fosfomycin, it is to be again reiterated that fosfomycin should not be used for empirical treatment of UTI as CLSI recommends it only for urinary E. coli and urinary E. faecalis [6]. In all 77% isolates were susceptible to nitrofurantoin [Table/Fig-3]. A susceptibility of 81.9% and 92.5% of urinary enterobacteriaceae isolates for nitrofurantoin was reported by Shevade G and Agrawal G and also by Ghosh AN et al., respectively [14,24]. In the class of non-fermenters, 55% and 40% isolates of P. aeruginosa and A. baumannii respectively were sensitive to piperacillin-tazobactam [Table/Fig-4]. Though their sensitivity to carbapenem was comparable, but carbapenems being reserved antimicrobials should not be used empirically in UTI. Shevade G and Agrawal G observed 68.7% sensitivity of non-fermenters causing nosocomial UTI for piperacillin-tazobactam and carbapenems [14].
WHONET surveillance data analysis for antimicrobial susceptibility of uropathogens in this study has shown that empirical treatment if given with nitrofurantoin, 100% UTI infections with staphylococci, 82% infections with E. faecalis and 77% infections with Enterobacteriaceae will be taken care of. As many as 55% infections with P. aeruginosa and 40% infections with A. baumannii will be taken care of by piperacillin-tazobactam. As piperacillin-tazobactam is to be given parenterally, its empirical use should be based on clinical assessment of the patient.
Mengistu A et al., in their study of empirical treatment of UTI at Namibia in Africa recommended the substitution of naldixic acid with fosfomycin as a first line treatment of community acquired UTI and continue the use of cefuroxime as second line as in the Namibia Standard Treatment Guidelines [25]. However, in the present study fosfomycin has not been recommended as an empirical drug as explained earlier. High resistance was encountered with cefuroxime in the present study [Table/Fig-2], hence not suitable for empirical therapy. Mengistu A et al., have reported high sensitivity of E. coli to nitrofurantoin, which is in agreement with the present study, but in contrast to the present study they found second common aetiological agent as P. mirabilis which showed high resistance to nitrofurantoin [25]. Mehrishi P et al., in their recent study at Himachal Pradesh observed that piperacillin-tazobactam and nitrofurantoin were the most sensitive antibiotics for the empirical therapy of UTIs [26]. Jayatilleke S et al., also observed high resistance rates in coliforms for orally available antibiotics with the exception of nitrofurantoin [27].
Department-wise comparison of aetiological agents of UTI and their antimicrobial susceptibility did not show significant difference (p>0.05) as shown in [Table/Fig-1]. Hence, a single hospital policy for empirical treatment of uropathogens is likely to be effective in all departments. The same database has been used for analysis of drug susceptibility of uropathogens by other researchers also [24,28-30]. The above facts reiterate the usefulness of WHONET in antimicrobial surveillance in a healthcare facility. Using this software we have found nitrofurantoin to be suitable for empirical treatment of UTI.
Limitation(s)
Categorisation by different age groups, history of prior antimicrobial therapy, duration of hospitalisation, clinical diagnosis, history of procedures like catheterisation were not taken into consideration being a retrospective laboratory based analysis of data.
Conclusion(s)
Management and analysis of microbiology data, especially the analysis of antimicrobial susceptibility test results becomes easier with the application of WHONET software. It provides a uniform and a standardised platform for a strong antimicrobial sensitivity surveillance that can guide in the formulation of antibiotic policy for the hospital. Nitrofurantoin was found to be the drug of choice for empirical treatment of UTI from this study. However, in serious hospitalised patients with UTI, additional parenteral administration of piperacillin-tazobactam may be considered. Empirically chosen antimicrobial treatment of uropathogens based on WHONET data analysis and subsequent correction based on antimicrobial susceptibility test report can improve patient care. It can further reduce complications, shorten hospital stay and help in financial savings.
1. Abbreviations and classes of uropathogens is as per the WHONET version 5.6.
2. @Gram positive cocci, #Enterobacteriaceae, $Non-fermenting gram negative bacilli, *Only 3 isolates of Pr mirabilis accounted for <1%, φOthers include Cardiovascular Thoracic Surgery, Endocrinology, Gastroenterology, Infectious Diseases, Burn, Neurology, Ophthalmology, Orthopaedics, Paediatric Surgery, Radiotherapy, Respiratory Medicine, Skin and Venereal Disease, Trauma Care Centre departments.
Percentage values are rounded off and not given in decimals. Little adjustments are made to get total as 100%.
4. By statistical test, no. of isolates of microbial groups in different departments are compared.
1. Percentage susceptibility of Proteus mirabilis and Citrobacter koseri has not been given, as the no. of isolates are very few. 2. *Fosfomycin tested for E coli only, 627 strains of E coli are susceptible from its total 639 isolates. 3. By statistical test, for different antibiotics susceptibility of E coli and K pneumoniae is compared.