TB mainly caused by MTB in humans, is a global health concern as more than 10 million new cases and 1.5 million deaths were reported in year 2018 [1]. While in developed countries like Japan, United States and Australia, the incidence rates of PNTM is higher than TB [2]. Both MTB and Non-Tuberculous Mycobacteria (NTM) usually cause chronic lung disease and could not be distinguished via clinical symptoms [3]. Previous TB infection will also increase the risk for NTM infection [3].
In low-resource settings, direct acid-fast sputum microscopy is the most widely used method to diagnose TB, but it has low sensitivity and specificity [4]. Thus, several detection kits with high sensitivity and specificity have been developed and endorsed by WHO for TB diagnosis such as Xpert MTB/RIF [5], Loopamp™ MTB Complex (MTBC) Detection Kit (Eiken Chemical Co., Japan) [6], and GenoType MTBDRplus (Hain Lifescience GmbH, Germany) [7]. The diagnosis of NTM is difficult as more than 180 different NTM has been reported [8]. Several tests are used for rapid differentiation of MTBC and NTM such as NTM+MDRTB Detection Kit 2 (Nipro Co., Japan) [9], GenoType Mycobacterium Common Mycobacteria/Additional Species (CM/AS) (Hain Lifescience, Germany) [10], DR. TBDR/NTM IVD kit (DR. Chip Corporation, Taiwan) [11], and CapitalBio Mycobacteria Real-Time PCR Detection Kit (CapitalBio Corporation, China). Several target genes such as hsp65, rpoB and 16S rRNA have been used for mycobacteria identification [12-14].
Despite of these antecedents, here, authors have reported the possibility to have false positive results with hsp65-PCR due to the identification of Rothia mucilaginosa caused by the ambiguity of the Tb11 and Tb12 hsp65 primers used.
Materials and Methods
Sample
In the context of a prospective study, a sputum sample from a 58-year-old, male with TB-like symptoms (fever and chronic cough) from a local community in Kota Kinabalu, Malaysia was collected. The study was approved by the Ethical Committee (Human) of Universiti Malaysia Sabah (UMS), in accordance with Declaration of Helsinki {JKEtika 2/2016(6)}. The ethical clearance for the TB screening program was obtained in June 2016, approved for three years of study. Informed consent was obtained from the patient.
Xpert MTB/RIF assay
Sputum (1 mL) was processed according to the manufacturer’s protocol by mixing with sample reagent (2 mL) in a 15 mL falcon tube and incubated at room temperature for 15 minutes. Then, the inactivated material (2 mL) was transferred to a test cartridge, loaded into Xpert MTB/RIF machine (Cepheid, Sunnyvale, CA) and the presence of MTB was determined by real-time PCR assay [18].
Sputum Processing and DNA Extraction
Sputum (1 mL) was decontaminated and digested with BBL® MycoPrep™ (Becton Dickinson, USA) (1 mL) for 15 minutes in a 50 mL falcon tube. The tube was top-up with Phosphate Buffered Saline (PBS) and the bacteria was pelleted by centrifugation at 3,000 g for 20 minutes. Bacterial DNA was extracted using Kaneka Easy DNA Extraction Kit version 2 (Konishiyasu Co., Japan), according to the manufacturer’s protocol. Solution A (100 μL) was added to the bacterial pellet and was incubated at 98°C for 8 minutes. Then, solution B (14 μL) was added to stop the chemical lysis. The DNA was stored at -20°C [19].
High-Fidelity hsp65-PCR and Sequencing
The hsp65-PCR assay was carried out in a total reaction volume of 50 μL with 5X PrimeSTAR GXL Buffer, 0.2 mM Deoxyribonucleoside Triphosphates (dNTPs), 200 nM Tb11 forward primer, 200 nM Tb12 reverse primer, and 1.25 U PrimeSTAR GXL DNA Polymerase (Takara Bio Inc., Japan) [20]. The DNA oligonucleotides, Tb11 (5’-ACCAACGATGGTGTGTCCAT) and Tb12 (5’-CTTGTCGAACCGCATACCCT) primers [16], were synthesised by Integrated DNA Technologies (IDT), Pvt., Ltd., Singapore. The extracted DNA (20 μL) was added to the master mix and amplified using the following thermal-cycling parameters: 1 cycle (95°C/2 min); followed by 45 cycles (98°C/10 sec, 60°C/15 sec and 68°C/1 min); and a final extension (68°C/5 min). DNA (1 μg/μL) from MTB H37Rv, Mycobacterium bovis BCG Pasteur strain TMC 1011, Mycobacterium avium subsp. paratuberculosis strain K-10, Mycobacterium avium subsp. avium strain 2285, Mycobacterium intracellulare strain 1956, Mycobacterium kansasii strain 824, Mycobacteriumsimiae strain MO-323 and Mycobacterium abscessus strain MA 1948, obtained from ATCC and BEI Resources, USA, were used as positive controls.
The amplified PCR products were analysed on a 2% agarose gel containing FloroSafe DNA stain (1st BASE, Singapore) and visualised using Molecular Imager® Gel Doc™ (Bio-Rad, USA).
The amplified PCR product from the patient was excised from the gel and purified using FavorPrep GEL/PCR Purification Kit (FAVORGEN Biotech Corp, Taiwan) according to the manufacturer instructions and sent for sequencing with Tb11 and Tb12 primers (Apical Scientific Sdn. Bhd., Malaysia).
Gene Alignment and Bioinformatic Analysis
The forward and reverse gene sequences obtained from sequencing were aligned using BioEdit software [21] and the National Center for Biotechnology (NCBI) Basic Local Alignment Search Tool (BLAST) software under blastn program [22] was used for bacterial identification. The sequence similarity between Tb11 and Tb12 primers with the identified bacteria was studied to determine the specificity of the primers using Primer-BLAST [23].
Results
Sputum Analysis with Xpert MTB/RIF and hsp65-PCR
The study of the patient’s sputum with Xpert MTB/RIF assay showed a negative result, suggesting non-Mycobacterium Tuberculosis Complex (MTBC) infection.
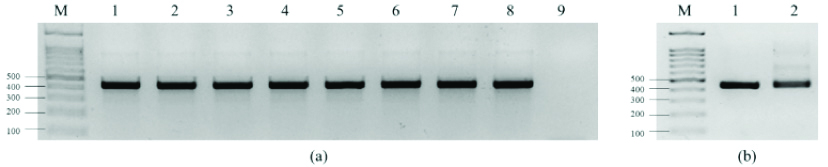
Tb11 and Tb12 primers specifically amplified the reference MTB and Non-Tuberculous Mycobacterial (NTM) DNA with the detection of a 441 bp hsp65 gene fragment [Table/Fig-1a]. Analysis of the DNA extracted from patient’s sputum also showed a positive amplification with similar characteristic [Table/Fig-1b], suggesting NTM infection or an Xpert MTB/RIF false negative result.
a) Amplification of DNA from Mycobacterium reference strains with Tb11 and Tb12 primers showed positive amplification with band size of 441 bp. M=DNA markers, 1=MTB H37Rv, 2=Mycobacterium bovis BCG Pasteur strain TMC 1011, 3=Mycobacterium avium subsp. paratuberculosis strain K-10, 4=Mycobacterium avium subsp. avium strain 2285, 5=Mycobacterium intracellulare strain 1956, 6=Mycobacterium kansasii strain 824, 7=Mycobacterium simiae strain MO-323, 8=Mycobacterium abscessus strain MA 1948, 9=Negative control; b) Amplification of DNA extracted from patient’s sputum with Tb11 and Tb12 primers showed positive amplification with same band size with MTB H37Rv. M=DNA markers, 1=MTB H37Rv, 2=Patient’s sputum.
Bioinformatic Analysis of the Amplified PCR Product from Patient
The DNA sequence alignment of the amplified PCR product from patient with BioEdit showed that the amplified gene fragment had 438 bp.
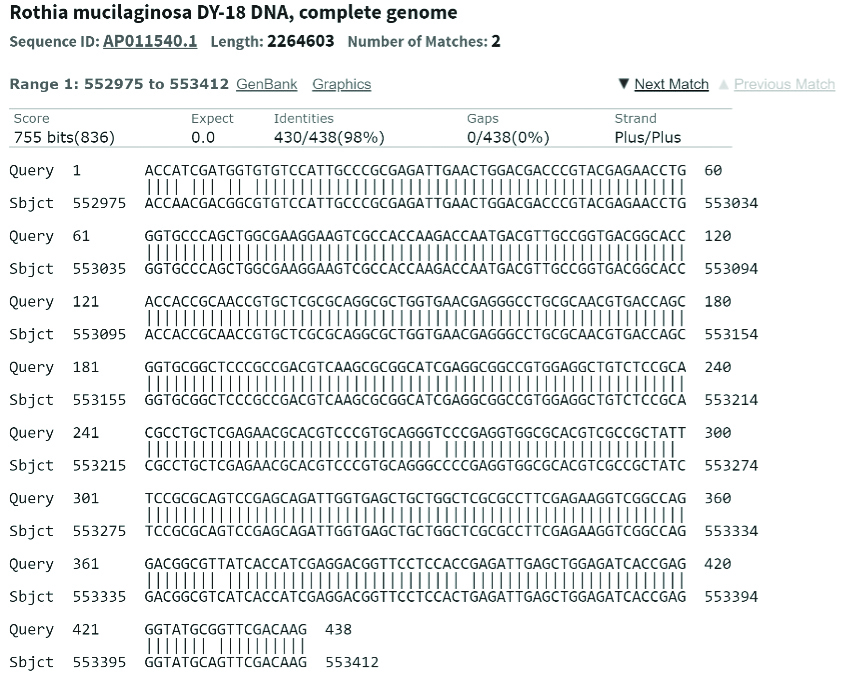
The bioinformatic analysis with blastn showed that the amplified product was closely related to Rothia mucilaginosa DY-18, with 100% query match and 98.17% of identity with eight nucleotide mismatches and E-value (expected value) of 0.0 [Table/Fig-2].
Analysis of the 438 bp gene fragment showing 100% query match and 98% similarity with Rothia mucilaginosa DY-18 whole genome (Image extracted from NCBI BLAST).
The top 20 sequences producing significant alignments with the hsp65-PCR amplified sequence in this study has been shown in [Table/Fig-3].
Top 20 sequences producing significant alignments with the hsp65-PCR amplified sequence in this study (438bp) (Data extracted from NCBI BLAST).
| # | Description | Max score | Total score | Query cover (%) | E-value | Per. ident (%) | Accession |
|---|
| 1 | Rothia mucilaginosa DY-18 DNA, complete genome | 755 | 951 | 100 | 0.0 | 98.17 | AP011540.1 |
| 2 | Rothia mucilaginosa strain FDAARGOS_369 chromosome, complete genome | 628 | 802 | 100 | 6e-176 | 91.78 | CP023510.1 |
| 3 | Rothia mucilaginosa DNA, complete genome, strain: NUM-Rm6536 | 628 | 780 | 100 | 6e-176 | 91.78 | AP014938.1 |
| 4 | Kocuria sp. BT304 chromosome, complete genome | 444 | 661 | 100 | 5e-120 | 82.42 | CP030039.1 |
| 5 | Kocuria rhizophila strain NCTC8340 genome assembly, chromosome: 1 | 439 | 666 | 100 | 6e-119 | 82.19 | LR134409.1 |
| 6 | Kocuria rhizophila strain FDAARGOS_302 chromosome, complete genome | 439 | 666 | 100 | 6e-119 | 82.19 | CP022039.2 |
| 7 | Kocuria rhizophila DC2201 DNA, complete genome | 439 | 666 | 100 | 6e-119 | 82.19 | AP009152.1 |
| 8 | Kocuria indica strain CE7 chromosome, complete genome | 421 | 643 | 100 | 2e-113 | 81.28 | CP035504.1 |
| 9 | Kocuria sp. KD4 chromosome, complete genome | 416 | 625 | 100 | 7e-112 | 81.05 | CP050449.1 |
| 10 | Arthrobacter sp. 11W110_air genome assembly PRJEB5507_assembly_1, scaffold CONTIG000002 | 416 | 416 | 100 | 7e-112 | 81.05 | LN483071.1 |
| 11 | Sinomonas atrocyanea strain KCTC 3377, complete genome | 412 | 631 | 100 | 8e-111 | 81.06 | CP014518.1 |
| 12 | Kocuria rosea strain ATCC 186 chromosome, complete genome | 411 | 613 | 100 | 3e-110 | 81.09 | CP035103.1 |
| 13 | Kocuria rosea strain NCTC7512 genome assembly, chromosome: 1 | 411 | 613 | 100 | 3e-110 | 81.09 | LR134487.1 |
| 14 | Kocuria rosea strain NCTC7514 genome assembly, chromosome: 1 | 411 | 613 | 100 | 3e-110 | 81.09 | LR134391.1 |
| 15 | Rothia dentocariosa strain NCTC10918 genome assembly, chromosome: 1 | 410 | 573 | 100 | 3e-110 | 81.02 | LR134521.1 |
| 16 | Rothia aeria DNA, complete genome | 407 | 575 | 100 | 3e-109 | 80.83 | AP017895.1 |
| 17 | Rothia dentocariosa strain NCTC10207 genome assembly, chromosome: 1 | 406 | 579 | 100 | 1e-108 | 80.79 | LR134479.1 |
| 18 | Kocuria flava strain HO-9041, complete genome | 397 | 597 | 100 | 6e-106 | 80.68 | CP013254.1 |
| 19 | Rothia dentocariosa ATCC 17931, complete genome | 397 | 560 | 100 | 6e-106 | 80.32 | CP002280.1 |
| 20 | Arthrobacter sp. QXT-31, complete genome | 385 | 600 | 100 | 1e-102 | 79.45 | CP019304.1 |
Per: ident-percentage identification
The analysis of the primers’ specificities with Rothia mucilaginosa DY-18 showed that the 20 bp TB11 primer has 18 similar nucleotides and two mismatches, while the reverse Tb12 primer has 19 similar nucleotides with one mismatch, based on the alignment of the Tb11 (forward primer) and Tb12 (reverse primer) with the Rothia mucilaginosa DY-18 whole genome DNA sequence (template) [Table/Fig-4]. According to the results of molecular study, the strain identified was Rothia mucilaginosa. Since this case was regarded as non-mycobacterial infections, the patient was not treated at specialised TB clinic and was referred to a nearby general health clinic for further management.
Tb11 and Tb12 primer sequence similarity with Rothia mucilaginosa DY-18 complete genome (Image extracted from Primer-BLAST).

Discussion
Diagnosis of TB via conventional sputum smear microscopy has low sensitivity, compared to culture and molecular methods [24]. Molecular detection of mycobacteria from direct clinical specimens has significantly reduced the time of detection [24]. Tb11 and Tb12 primers have been commonly used for direct hsp65 gene amplification in clinical samples and pure culture for the detection of mycobacteria [17,25,26]. However, in this study, Rothia mucilaginosa was identified using hsp65-PCR diagnostic method, showing a false positive identification, which could wrongly be interpreted as an NTM infection or an Xpert MTB/RIF false negative result.
The analysis of the top 20 sequences producing significant alignments with the hsp65-PCR amplified sequence in present study showed that Rothia mucilaginosa DY-18 had the highest identity and lowest E-value, which supported the identification of this strain with high degree of confidence. The E value obtained for Rothia mucilaginosa DY-18 (0.0) means that zero sequences can be expected to match better [27].
Rothia mucilaginosa, is part of the normal flora residing in the human oropharynx and upper respiratory tract [28]. It is recognised as opportunistic pathogen in immuno-compromised individuals, resulting in pneumonia [28].
The non-specific amplification obtained with the hsp65-PCR assay used can be explained by high similarity of the primers used with the Rothia mucilaginosa gene sequence, with few mismatches to the middle of the primers. It is considered that a target can be amplified even if there are few mismatches in the middle or towards the 5’ end of the primers sequence [23]. According to the general rules in specific primer design, two base mismatches at the 3’ end are generally required to prevent non-specific amplification [23].
A study by Randima GDD et al., to detect mycobacteria in elephant nasal secretions collected from trunk wash samples, using the same set of primers of present study, showed poor sensitivity in mycobacteria detection and DNA sequencing of the amplified products showed the best match with Rothia dentocariosa [29].
Busatto C et al., used hsp65 as a marker (primer sequences not disclosed) for detection of Mycobacterium avium in suspected-TB patients’ DNA samples that were IS6110-PCR negative. The sequencing results of positive hsp65 amplifications showed the presence of not only mycobacteria (MTBC, Mycobacterium avium and Mycobacteriummonacense), but also other organisms such as Rhodococcus spp., Rothia mucilaginosa, Gordonia spp., Cryobacterium spp., Streptomyces spp., Nocardia spp., and Corynebacterium spp., suggesting the low accuracy of the diagnostic technique [30].
Sarti E et al., using the same primers, reported the amplification of Bifidobacterium crudilactis, Kocuria rhizophila and Kocuria palustris in cheese samples [31]. The presence of Kocuria spp., had been reported in the skin and mucosa of animals and human [32], and this could be another source of false positive result when hsp65-PCR is used for identification of mycobacteria.
Kim BJ et al., using different primers, designed a hsp65 nested PCR-direct sequencing method to increase the sensitivity for species identification from sputum, which according to the primers design avoid Rothia spp. amplification [33]. The use of this method could be an alternative to eliminate the possibility of Rothia spp. amplification.
Limitation(s)
The culture method (gold standard) for detection of mycobacteria either on Löwenstein-Jensen medium or in Mycobacteria Growth Indicator Tube (MGIT) was not used in this study due to limited sample availability.
Conclusion(s)
The present study showed the possibility of false positive results in clinical specimens using PCR-hsp65 primers considered specific for mycobacteria, therefore, this test should be used in clinical samples with caution and it is suggested the need of its further re-optimisation.