Evaluation of Bacteriological Profile and their Antimicrobial Susceptibility Pattern in Blood Stream Infections in a Multispeciality Hospital
Mariyah Yousuf1, Tarana Sarwat2, Dalip K Kakru3
1 PG Scholar (MSc), Department of Microbiology, School of Medical Science and Research, Sharda University, Greater Noida, Uttar Pradesh, India.
2 Assistant Professor, Department of Microbiology, School of Medical Science and Research, Sharda University, Greater Noida, Uttar Pradesh, India.
3 Professor, Department of Microbiology, School of Medical Science and Research, Sharda University, Greater Noida, Uttar Pradesh, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Miss. Mariyah Yousuf, Department of Microbiology, School of Medical Science and Research, Sharda University, Knowledge Park 3, Greater Noida, Uttar Pradesh, India.
E-mail: mariasauban20@gmail.com
Introduction
Bloodstream Infections (BSIs) are one of the major cause of morbidity and mortality worldwide. To decrease the mortality from septicaemia early diagnosis and appropriate treatment of BSIs is most important. Early diagnosis of a BSI will markedly improve patient management.
Aim
To identify various pathogenic organisms causing BSIs and determine their susceptibility to various antibiotics.
Materials and Methods
A total of 1367 blood samples were received in the bacteriology laboratory for culture out of which 274 samples showed culture positivity. Blood cultures were repeated for confirmation of results. Simple descriptive analysis of data was done and results presented in frequencies and percentages.
Results
Out of 274 positive samples obtained, Coagulase Negative Staphylococci (CoNS) constituted maximum proportion of isolates (66%) followed by Pseudomonas species (12%), Escherachia coli (6.2%), Klebsiella species (3.2%), Citrobacter spp. (2.9%), Staphylococcus aureus (2.9%) and Enterococcus (2.9%). Among the positive cases, 32 cases of Multidrug Resistance (MDR) were found. MDR cases show resistance to ≥3 classes of antibiotics. Enterobacteriaceae family showed highest MDR cases.
Conclusion
The present study highlighted the bacteriological aetiology of BSIs along with their antibiogram that may provide necessary information for the formulation of antibiotic policy in effective management of such cases.
Bacteremia, Blood culture, Coagulase negative staphylococcus, Multidrug resistance, Septicaemia
Introduction
Approximately 200,000 cases of bacteraemia and fungemia occur annually with mortality rates ranging from 20-50% [1]. Blood cultures in which contamination has been effectively ruled out and viable bacteria are observed in it, then bacteremia is considered [2,3]. The most common bacteria that cause bacteremia include members of Staphylococcus spp., Streptococcus spp., Enterococcus spp., Escherichia coli, Klebsiella spp., Pseudomonas spp., Enterobacter spp., Haemophilus spp., and Neisseria genera [4-6]. Septicaemia can lead to serious complications such as shock, disseminated intravascular coagulation, multiple organ failure, etc. Thus, BSIs are one of most serious pathologies, so early detection and identification of blood stream pathogen is important [7]. Provisional diagnosis of septicaemia can be carried out by clinical assessment using a combination of symptoms and signs. But, identifying the causative pathogen by bacteriologic culture is necessary for definitive diagnosis of septicaemia [8]. Early diagnosis of bacteremia can be obtained by blood culture and antimicrobial susceptibility test which helps in identifying the most appropriate effective antibiotic which can be a choice of drug to be administered and thus helps in early recovery and reducing mortality due to septicaemia. This can reduce turnaround time and improve patient management.
In a particular area knowing the epidemiology and antibiotic susceptibility pattern of a various pathogenic organisms will help to determine the antibiotic of choice and thus assist in proper management. This study will identify the most common pathogens causing BSIs and will provide their antimicrobial susceptibility pattern.
Materials and Methods
This retrospective study was conducted in the Department of Microbiology, Central Laboratory, School of Medical Sciences and Research, Sharda Hospital, Greater Noida, Uttar Pradesh, India. This study was conducted for a period of six months from May 2019 to October 2019. Institutional Ethics Committee provided approval for this study (Ref no. - SU/SMS&R/76-A/2019/35). All the isolates from blood culture positive samples collected over period of six months were included in the study. All the blood samples received in bacteriological laboratory for culture were used for study. Automated method i.e., (BACT-ALERT system) was used to culture the bottles received in the bacteriology laboratory. The bottles were put inside the automated machine, when there was a signal the bottles were removed and the blood from the positive blood culture bottles was subjected to subculture on 5% sheep blood agar and MacConkey agar. The growth on culture plates was identified on the basis of colony morphology, Gram stain and various biochemical tests. Blood cultures were repeated for confirmation of results. Antimicrobial susceptibility testing was performed for all blood cultures isolated on Muller Hinton agar by Kirby-Bauer disc diffusion method as recommended in the CLSI (Clinical and Laboratory Standards Institute guidelines) 2019. Commercially, available antibiotics disks (Himedia) were used for antimicrobial susceptibility testing [Table/Fig-1]. The antibiotics used for Gram positive bacteria were levofloxacin (5 ug), clindamycin (2 ug), ciprofloxacin (5 ug), linezolid (30 ug), penicillin (10 units), erythromycin (15 ug), gentamicin (10 ug), vancomycin, cefotaxime (30 ug) and the antibiotics used for gram negative bacteria were cefotaxime (30 ug), cefepime (30 ug), cefuroxime (30 ug), levofloxacin (5 ug), ampicillin (10 ug), gentamicin (10 ug), imipenem (10 ug), meropenem (10 ug), amoxiclav (30 ug), ceftriaxone (30 ug).
Antibiotic susceptibility test (disk diffusion method).
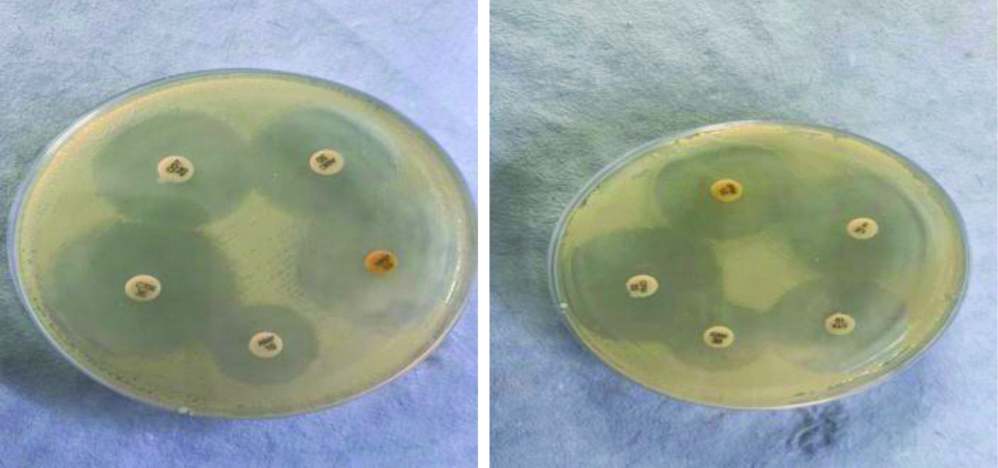
Statistical Analysis
Simple descriptive analysis of the distribution of sample, age, gender, and antimicrobial susceptibility data were done, and the results were obtained and presented as frequencies and percentages.
Results
A total of 1367 blood samples were received in the bacteriology lab for culture, out of which 274 samples showed culture positivity. Out of 274 positive samples obtained, 156 were male patients and 118 were females. The maximum number of samples was obtained from patients between age group 0-10 years followed by the age group 40-50 and 60-70 years [Table/Fig-2].
Age and sex wise distribution of samples.
| Age group (Years) | Males | Females | Total |
|---|
| 0-10 | 79 | 59 | 138 |
| 10-20 | 09 | 10 | 19 |
| 20-30 | 14 | 05 | 19 |
| 30-40 | 08 | 07 | 15 |
| 40-50 | 16 | 08 | 24 |
| 50-60 | 07 | 15 | 22 |
| 60-70 | 17 | 07 | 24 |
| 70-80 | 06 | 06 | 12 |
| 80-90 | 0 | 01 | 01 |
| Total | 156 | 118 | 274 |
CoNS constituted maximum proportion of isolates followed by Pseudomonas species and E. coli. [Table/Fig-3]. Most of the cases were positive for coagulase negative organisms and amongst all the antibiotics, vancomycin (100%) emerged as most sensitive drug against CoNS followed by linezolid (90%), and clindamycin (55%). Strains of CoNS showed high resistance for erythromycin (76.2%) followed by penicillin (65.7%) and ciprofloxacin (59.6%) [Table/Fig-4].
Distribution of organisms in positive samples.
| Organisms | Number | % |
|---|
| Coagulase negative staphylococcus | 181 | 66.0 |
| Staphylococcus aureus | 08 | 2.9 |
| Enterococcus | 08 | 2.9 |
| Escherichia coli | 17 | 6.2 |
| Klebsiella spp. | 09 | 3.2 |
| Citrobacter spp. | 08 | 2.9 |
| Pseudomonas spp. | 33 | 12.0 |
| Acinetobacter spp. | 07 | 2.5 |
| Sphingomonas spp. | 02 | 0.72 |
| Burkholderia cepacia | 01 | 0.36 |
Antibiotic susceptibility pattern of coagulase negative Staphylococcus (CoNS) (n=181).
| Antibiotics | (Number of sensitive samples) | Sensitive % | Number of resistant samples | Resistant % |
|---|
| Vancomycin | 181 | 100% | 0 | 0 |
| Linezolid | 164 | 90% | 17 | 9.3% |
| Gentamicin | 139 | 77% | 42 | 23% |
| Ciprofloxacin | 73 | 40% | 108 | 59% |
| Levofloxacin | 99 | 54% | 82 | 45% |
| Erythromycin | 43 | 23% | 138 | 76% |
| Penicillin | 62 | 34% | 119 | 65% |
| Clindamycin | 101 | 55% | 80 | 44% |
| Cefotaxime | 85 | 46% | 96 | 53% |
Amongst all the antibiotics, linezolid and vancomycin emerged as most sensitive drug against Staphylococcus spp. Strains of Staphylococcus spp. showed high resistance for Penicillin. Linezolid was found most sensitive drug against Enterococcus spp. Strains Enterococcus spp. showed high resistance for ampicillin and penicillin. E.coli was found most susceptible to gentamicin, imipenem, cotrimoxazol and meropenem. Strains of Escherichia coli showed high resistance for cefuroxime and ceftriaxone [Table/Fig-5].
Antibiotic susceptibility pattern of Escherichia coli species.
| Antibiotics | Sensitive N% | Resistant N% |
|---|
| Ampicillin | 08 (47%) | 09 (53%) |
| Amoxyclav | 09 (53%) | 08 (47%) |
| Cefepime | 13 (76%) | 04 (24%) |
| Cefotaxime | 08 (47%) | 09 (53%) |
| Ceftriaxone | 06 (35%) | 11 (65%) |
| Cefuroxime | 06 (35%) | 11 (65%) |
| Gentamicin | 16 (94%) | 01 (6%) |
| Imipenem | 14 (82%) | 03 (18%) |
| Levofloxacin | 11 (65%) | 06 (35%) |
| Meropenem | 13 (76%) | 04 (08%) |
| Cotrimoxazole | 14 (82%) | 03 (18%) |
Imipenem (89%) and meropenem (77%) emerged as most sensitive drug against Klebsiella spp. Strains of Klebsiella spp. showed high resistance for cefepime (89%), ceftrioxone and ampicillin (78%). Ampicillin and gentamicin were found most effective against Citrobacter spp. Strains of Citrobacter spp. showed high resistance for cefepime. Piperacillin+Tazobactum and Levofloxacin were most efficient against Pseudomonas spp.
Strains of Pseudomonas spp. showed high resistance for Aztreonam [Table/Fig-6]. Gentamicin and Levofloxacin were observed most sensitive drug against Acinetobacter spp. Strains of Acinetobacter spp. showed high resistance for tetracycline and ceftriaxone.
Antibiotics suscetivility pattern in Pseudomonas spp.
| Antibiotics | Sensitive N% | Resistant N% |
|---|
| Aztreonam | 12 (37%) | 21 (63%) |
| Ceftazidime | 16 (48%) | 17 (51%) |
| Ciprofloxacin | 25 (75%) | 08 (24%) |
| Levofloxacin | 29 (87%) | 04 (12%) |
| Meropenem | 25 (75%) | 08 (25%) |
| Piperacillin | 25 (75%) | 08 (24%) |
| Piperacillin-Tazobactum | 29 (87%) | 04 (12%) |
| Tobramycin | 22 (66%) | 11 (33%) |
| Ticarcillin | 21 (63%) | 12 (37%) |
MDR was observed in approximately 32 positive culture isolates which constituted 11.6% of total positive cultures. Gram negative bacilli constituted higher percentage (27, 10%) than gram positive organism (5, 1.8%) among MDR isolates. Among gram negative organisms Pseudomonas spp. were most common isolates to demonstrate MDR [Table/Fig-7].
Distribution of multi drug resistant organism.
| Organisms | Number | Percentage |
|---|
| CoNS | 0 | 100 |
| Citrobacter spp. | 4 | 12.5 |
| Klebsiella spp. | 4 | 12.5 |
| Pseudomonas spp. | 9 | 28 |
| E.coli | 6 | 18 |
| Citrobacter spp. | 2 | 6.2 |
| Acinetobacter spp. | 2 | 6.2 |
| Staphylococcus spp. | 3 | 9 |
| Enterococcus spp. | 2 | 6.2 |
Discussion
In this study, gram positive bacteria were more common isolates than gram negative organism. Gram positive bacteria accounted for 71% of the isolates. This was also observed by Prabhu K et al., who found 64% of isolates were gram positive organisms [9]. Among gram positive bacteria CoNS was isolated in most samples (92%) than Staphylococcus aureus (4%). Jamal WY et al., found that most common isolate in BSI was CoNS (46%) [10]. In a similar study done by Valles J et al., CoNS accounted for 49.8% of the isolates [11]. Wattal C et al., reported CoNS as the most common isolate causing BSIs in ICU patients [12]. It has been demonstrated by some studies upto 85% of CoNS represent contamination rather than true bacteremia [13]. In order to avoid this repeat blood cultures were done for confirmation. In the present study, gram negative bacilli accounted for 28.1 of BSIs, similar observations were done by Prabhu K et al., who observed 35% of the culture isolates were gram negative bacilli [9]. Pseudomonas spp. are most common and accounted for 42.6% among gram negative organisms.
In this study, males were more commonly involved accounting for 56.9% of total number of positive cases and females account for approximately 43% of total number of positive cases. The result was consistent with the study done by Kaur A and Singh V who reported high culture positivity in men 65.2% [14]. Hussein A et al., reported 66.6% positivity in men and 33.3% in women [15]. Zenebe T et al., found that women (59.8%) were effected more than men (40.2%) [16].
From amongst the bacteria isolated in present study, CoNS showed 100% sensitivity to vancomycin and 90% sensitivity to linezolid. Vancomycin and Linezolid were sensitive to all species of CoNS in the study by Asangi SY et al., and Singh L et al., [17,18]. The organisms were found highly resistant to erythromycin (76%) followed by penicillin (65%). The second common isolate among gram positive bacteria was Staphylococcus spp. which showed 100% sensitivity to linezolid and vancomycin, followed by clindamycin (75%). The organisms were found highly resistant to penicillin (87%) followed by erythromycin (75%) and cefoxitin (62%). The antimicrobial susceptibility testing revealed that resistance to penicillin was frequent in Staphylococcus spp. (87%) and CoNS (65%) this was consistent with the study of Roy I et al., who observed 89% of Staphylococcus isolated were resistant to pencillin and none of isolates were resistant to vancomycin [19]. Among gram negative organisms, Pseudomonas spp. was the most common isolate in which Piperacillin+Tazobactum (87%) emerged as most sensitive drug followed by piperacillin (75%), meropenem (75%) and ciprofloxacin (75%). Patel PH et al., observed 100% susceptibility to imipenem of Pseudomonas spp. [20]. However, Pourakbari B et al., had reported resistance to most of antibiotics by Pseudomonas spp. Imipenem (89%) emerged as most sensitive drug against Klebsiella spp. followed by meropenem (77%) and cotrimoxazole (67%). Strains of Klebsiella showed high resistance for cefepime (89%) followed by ceftriaxone (78%), ampicillin (78%) and amoxyclav (55%). Citrobacter spp. was highly sensitive to imipenem (75%) and appear resistant to ampicillin (62%) [21]. Sphingomonas paucimobilis was detected in 2 samples. It was highly sensitive to imipenem (100%) which was consistant with study of Bayram N et al., [22]. Carbapenems were the most effective therapy. Burkholderia cepacia was found in single culture constituting a mere 0.36% of total number of isolates. Gautam V et al., isolated 39 isolates of Burkholderia cepacia from various specimens which accounted for 0.35% of total number of isolates [23]. It showed sensitivity to co-trimoxazole, imipenem, levofloxacin and appeared resistant to cephalosporins, ampicillin and gentamicin. In this study, 32 isolates showed MDR which constitute 11.6% of total number of positive cultures with gram negative organisms most commonly involved. Among gram negative organisms Pseudomonas spp. mainly show MDR. Tam VH et al., observed that in P. aeruginosa bloodstream isolates approximately 10-17% showed MDR [24]. The number of isolates obtained in this study has been compared with other previous studies [Table/Fig-8] [25-30].
Comparison of present study isolates with other Indian studies [25-30].
| Various studies/Year | CoNS | Staphylococcus spp. | E. coli | Klebsiella spp. | Acinetobacter spp | Pseudomonas spp. | Enterococcus | Burkholderia spp. |
|---|
| Present study | 66% | 2.9% | 6.2% | 3.2% | 2.5% | 12% | 2.9% | 0.3% |
| Mathur P et al., 2014 [25] | 4% | 14.5% | 4% | 18% | 21.5% | 8% | 1.5% | 2% |
| Gohel K et al., 2014 [26] | 4.5% | 38.6% | 15.2% | 9.8% | 1.5%1 | 5.3% | 3.8% | 4.5% |
| Singhal T et al., 2016 [27] | 8% | 8% | 26% | 22% | 10% | 8% | 8% | 1.2% |
| Prabash K et al., 2010 [28] | 10.5% | 12.6% | 10.9% | 4.5% | 11.5% | 30.3% | 4.1% | -- |
| Garg A et al., 2007 [29] | 20.7% | 8,3% | 11% | 7.3% | 12.6% | 16% | 3.7% | -- |
| Khurana S et al., 2018 [30] | 6.8% | 7.1% | 5.1% | 15.2% | 24.1% | 9.6% | 1.4% | 10.6% |
Thus, the present study clearly indicates that most common isolates from BSIs were CoNS which appeared highly sensitive to vancomycin and linezolid whereas, among gram negative organisms, Escherichia coli were commonly isolated. MDR isolates also constituted a significant number and were commonly found among gram negative organisms.
Limitation(s)
This study was conducted for shorter period of time that is six months and thus chances for identifying risk factors for mortality were reduced. Another limitation of this study was its retrospective analysis which may be prone to selection bias which was omitted by selecting all samples in present study.
Conclusion(s)
This study formed a useful reference for clinical microbiologists, physicians and others attempting to monitor the prevalence of BSIs and for the treatment of patients with such infections. Utilising various methods for reduction of development of antibiotic resistance is utmost requirement which can be established by adopting specific antibiotic utilisation strategies e.g., reducing antibiotic usage, developing combination therapy, using antibiotics only after standard antimicrobial susceptibility testing and recycling of antibiotics. Using methods for proper infection control and proper channelization of antibiotic programs are most important and are of prime requirement.
Author Declaration:
Financial or Other Competing Interests: None
Was Ethics Committee Approval obtained for this study? Yes
Was informed consent obtained from the subjects involved in the study? No
For any images presented appropriate consent has been obtained from the subjects. No
Plagiarism Checking Methods: [Jain H et al.]
Plagiarism X-checker: May 04, 2020
Manual Googling: Jun 29, 2020
iThenticate Software: Jul 31, 2020 (11%)
[1]. Karlowsky JA, Jones ME, Draghi DC, Thornsberry C, Sahm DF, Voltaire GA, Prevalance and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalised patients in the United States in 2002Annals of Clinical Microbiology and Antimicrobials 2004 3:710.1186/1476-0711-3-715134581 [Google Scholar] [CrossRef] [PubMed]
[2]. Kuppermann N, Occult bacteremia in young febrile childrenPediatr Clin North Am 1999 46(6):n1073-110.10.1016/S0031-3955(05)70176-0 [Google Scholar] [CrossRef]
[3]. Bryan C, Clinical implication of positive blood culture Clin Microbiol Rev 1989 2(4):329-53.10.1128/CMR.2.4.3292680055 [Google Scholar] [CrossRef] [PubMed]
[4]. Panceri M, Vegni F, Goglio A, Manisco A, Tambini R, Lizioli A, An etiology and prognosis of bacteremia in ItalyEpidermiol Infect 2004 132(4):647-54.10.1017/S095026880300108015310166 [Google Scholar] [CrossRef] [CrossRef]
[5]. Pedersen G, Schonheyder H, Sorensen Source of infection and other factors associated with case fatality in community acquired bacteremia- A Danish population based cohort study from 1992-1997Clin Microbiol Infect 2003 9(8):793-802.10.1046/j.1469-0691.2003.00599.x14616699 [Google Scholar] [CrossRef] [PubMed]
[6]. Ali H, Ali M, Moammar N, Zeana S, Ruqia M, Nebras M, Etiological agents of bacteremia among newbporns in Hilla cityMed J Babylon 2008 5(4):385-94. [Google Scholar]
[7]. Forbes BA, Sahm DF, Weissfeld AS, Bailey and Scott’s Diagnostic Microbiology 2007 MissouriMosby Elsevier:779 [Google Scholar]
[8]. Meremkwer MM, Nwachukwu CE, Asuquo AE, Okebe J, Utsalo SJ, Bacterial isolates from blood cultures of children with suspected septicaemia in Calabar, NigeriaBMC Infect Dis 2005 5:110-15.10.1186/1471-2334-5-11016336657 [Google Scholar] [CrossRef] [PubMed]
[9]. Prabhu K, Bhat S, Rao S, Bacteriologic profile and antibiogram of blood culture isolates in a pediatric care unitJ Lab Physicians 2010 2:85-88.10.4103/0974-2727.7215621346903 [Google Scholar] [CrossRef] [PubMed]
[10]. Jamal WY, El-Din K, Rotimi VO, Chugh TD, An analysis of hospital acquired bacteraemia in intensive care unit patients in a university hospital in KuwaitJ Hosp Infect 1999 43:49-56.10.1053/jhin.1999.060810462639 [Google Scholar] [CrossRef] [PubMed]
[11]. Valles J, Leon C, Alvarez-Lerma F, Nosocomial bacteremia in critically ill patients: A multicenter study evaluating epidemiology and prognosisClin Infect Dis 1997 24:387-95.10.1093/clinids/24.3.3879114190 [Google Scholar] [CrossRef] [PubMed]
[12]. Wattal C, Raveendran R, Goel N, Oberoi JK, Rao BK, Ecology of blood stream infection and antibiotic resistance in Intensive Care Unit at a tertiary care hospital in North IndiaBraz J Infect Dis 2014 18:245-51.10.1016/j.bjid.2013.07.01024389282 [Google Scholar] [CrossRef] [PubMed]
[13]. Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, The clinical significance of positive blood cultures in the 1990s: A prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adultsClin Infect Dis 1997 24:584-602.10.1093/clind/24.4.5849145732 [Google Scholar] [CrossRef] [PubMed]
[14]. Kaur A, Singh V, Bacterial isolates and their antibiotic sensitivity pattern in clinically suspected cases of fever of unknown originJK Science 2014 16:105-09. [Google Scholar]
[15]. Hussein A, Sayed A, Mohamed A, Seroepidemiological study on human brucellosis in Assiut GovernorateEgypt J Immunol 2005 12:49-56. [Google Scholar]
[16]. Zenebe T, Kannan S, Yilma D, Beyene G, Invasive bacterial pathogens and their antibiotic susceptibility patterns in Jimma University Specialized Hospital, Jimma, Southwest EthiopiaEthiop J Health Sci 2011 21:01-08.10.4314/ejhs.v21i1.6903822434980 [Google Scholar] [CrossRef] [PubMed]
[17]. Asangi SY, Mariraj J, Sathyanarayan MS, Nagabhushan R, Speciation of clinically significant coagulase negative staphylococci and their antibiotic resistant pattern in a tertiary care hospitalInt J Biol Med Res 2011 2:735-39.10.1016/j.mjafi.2016.07.00928050073 [Google Scholar] [CrossRef] [PubMed]
[18]. Singh L, Cariappa MP, Das NK, Drug sensitivity pattern of various Staphylococcus species isolated at a tertiary care hospitalMed J Armed Forces India 2016 72:S62-66. [Google Scholar]
[19]. Roy I, Jain A, Kumar M, Aggarwal SK, Bacteriology of neonatal septicaemia in a tertiary care hospital of northern IndiaIndian Journal of Medical Microbiology 2002 20(3):156-59.10.1016/S0255-0857(21)03250-3 [Google Scholar] [CrossRef]
[20]. Patel PH, Pethani JD, Rathod SD, Chauhan B, Shah PD, Prevalence of nonfermenting gram negative bacilli infection in tertiary care Hospital in Ahmedabad, GujaratIndian Journal of Basic and Applied Medical Research 2013 6(2):608-13. [Google Scholar]
[21]. Pourakbari B, Sadr A, Ashtiani MTH, Mamishi S, Dehghani M, Mahmoudi S, Five-year evaluation of the antimicrobial susceptibility patterns of bacteria causing bloodstream infections in IranJ Infect Dev Ctries 2012 6(2):120-25.10.3855/jidc.151722337839 [Google Scholar] [CrossRef] [PubMed]
[22]. Bayram N, Devrim I, Apa H, Gülfidan G, Türkyılmaz HN, Günay I, Sphingomonas paucimobilis infections in children: 24 case reportsMediterr J Hematol Infect Dis 2013 5(1):e201304010.4084/mjhid.2013.04023795278 [Google Scholar] [CrossRef] [PubMed]
[23]. Gautam V, Singhal L, Ray P, Burkholderia cepacia complex: Beyond pseudomonas and acinetobacterIndian J Med Microbiol 2011 29(1):04-12.10.4103/0255-0857.7651621304187 [Google Scholar] [CrossRef] [PubMed]
[24]. Tam VH, Chang KT, Abdelraouf K, Brioso CG, Ameka M, McCaskey LA, Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosaAntimicrob Agents Chemother 2010 54(3):1160-64.10.1128/AAC.01446-0920086165 [Google Scholar] [CrossRef] [PubMed]
[25]. Mathur P, Varghese P, Tak V, Gunjiyal J, Lalwani S, Kumar S, Epidemiology of blood stream infections at a level-1 trauma care center of IndiaJ Labs Physicians 2014 6:22-27.10.4103/0974-2727.12908624696556 [Google Scholar] [CrossRef] [PubMed]
[26]. Gohel K, Jojera A, Soni S, Gang S, Sabnis R, Desai M, Bacteriological profile and drug resistance patterns of blood culture isolates in a tertiary care nephrourology teaching instituteBiomed Res Int 2014 2014:15374710.1155/2014/15374724804199 [Google Scholar] [CrossRef] [PubMed]
[27]. Singhal T, Shah S, Naik R, The microbial etiology and antimicrobial susceptibility of bloodstream infections in patients with cancer at a private tertiary care hospital in Mumbai, IndiaIndian J Cancer 2016 53:452-53. [Google Scholar]
[28]. Prabash K, Medhekar A, Ghadyalpatil N, Noronha V, Biswas S, Kurkure P, Blood stream infections in cancer patients: A single center experience of isolates and sensitivity patternIndian J Cancer 2010 47:184-88.10.4103/0019-509X.6301920448384 [Google Scholar] [CrossRef] [PubMed]
[29]. Garg A, Anupurba S, Garg J, Bacteriological profile and antimicrobial resistance of blood culture isolates from a university hospitalJournal of Indian Academy of Clinical Medicine 2007 8(2):139-43. [Google Scholar]
[30]. Khurana S, Bhardwaj N, Kumari M, Malhotra R, Mathur P, Prevalence, etiology, and antibiotic resistance profiles of bacterial bloodstream infections in a tertiary care hospital in Northern India: A 4-year studyJ lab Physicians 2018 10(4):426-31.10.4103/JLP.JLP_78_1830498316 [Google Scholar] [CrossRef] [PubMed]