Materials and Methods
A retrospective study was conducted on the V. cholerae isolates from the stool samples during the period from June 2019 to December 2019 at the Department of Microbiology, Tertiary Care Hospital, Vadodara, Gujarat, India. Permission was taken from Ethics Committee (No. IECBHR-50/2020 dated 05/06/2020). During this study, all clinically suspected cases of cholera were included. As a routine procedure, informed consent from patients was obtained when sample was collected. Stool samples received in the laboratory were analysed and V. cholerae was isolated in 72 patients. Repeat isolates from the same patients were not included in this study.
Stool specimens were collected on admission in a clean, wide mouthed, leak proof container preferably before starting antibiotics and transported immediately to the laboratory for processing. Specimens were cultured directly on Nutrient agar, Blood agar, MacConkey’s agar, Thiosulfate-citrate-bile salts (TCBS) sucrose agar [Table/Fig-1]. Nutrient agar was used to perform biochemical tests on the isolates and Blood agar was used for biotyping. Further, the specimens were sub cultured on MacConkey’s agar, TCBS agar after enrichment in Alkaline peptone water and were examined after overnight incubation at 37°C [10].
a) Nutrient agar, b) Mac Conkey agar, c) Nutrient agar showing isolated colonies of Vibrio cholerae.
Colonies suggestive of V. cholerae were identified by standard biochemical tests. Serotyping of isolates was done for agglutination using V. cholerae polyvalent, V. cholerae Ogawa and V. cholerae Inaba antisera (Denka Seiken Co., Ltd., Japan). Biotyping was done by conventional Methods such as Polymyxin-B sensitivity and Sheep RBC haemolysis [10].
Phage typing was done by Basu S and Mukherjee S method and New Phage Typing method and susceptibility to phage IV and V for biotyping was done at NICED, Kolkata, West Bengal, India [7,8].
Antibiotic susceptibility testing was performed by Kirby Bauer’s Method as per Clinical and Laboratory Standards Institute (CLSI) guidelines [11]. The control used for antimicrobial sensitivity testing was ATCC strain Escherichia coli 25922. The following antibiotics were used-Ceftriaxone (30 μg), Doxycycline (30 μg), Levofloxacin (10 μg), Ampicillin (30 μg) and Cotrimoxazole (25 μg). The results were seen after 16-18 hours of incubation at 37°C. The zone of inhibition for each antibiotic was interpreted as per CLSI guidelines [11].
Statistical Analysis
Descriptive analysis was used and presented in terms of percentage.
Results
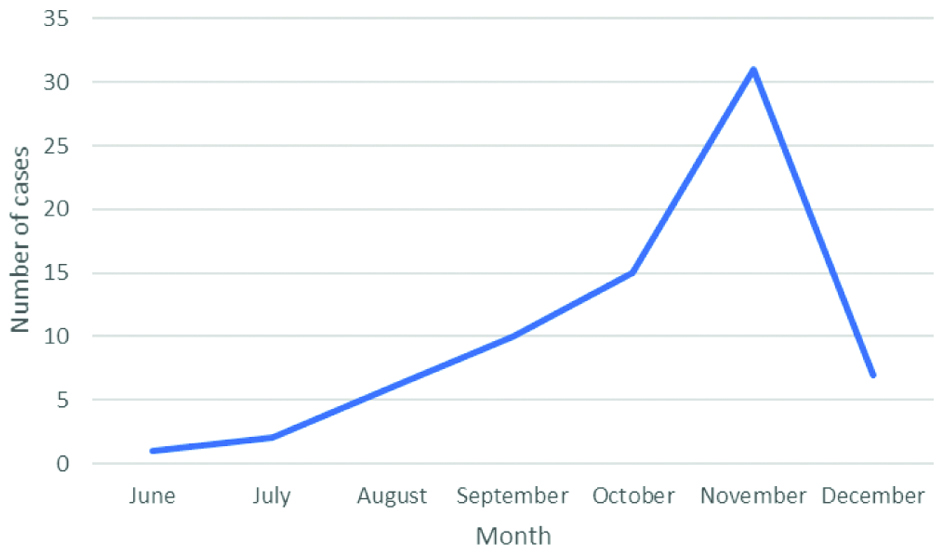
V. cholerae was isolated from 72 patients. Of the 72 patients, 53 (73.6%) were adults and 19 (26.4%) were children. Male to female ratio was 1.4:1. Maximum number of cases was in the month of November which were 32 isolates followed by 15 isolates in October [Table/Fig-2].
Seasonal pattern of occurrence of cases.
All the 72 isolates of V. cholerae belonged to serogroup O1, biotype El Tor and serotype Ogawa. None of the isolates belonged to serotype Inaba. No discordance was found for serotyping and biotyping done in the laboratory and results provided by NICED, Kolkata, West Bengal, India.
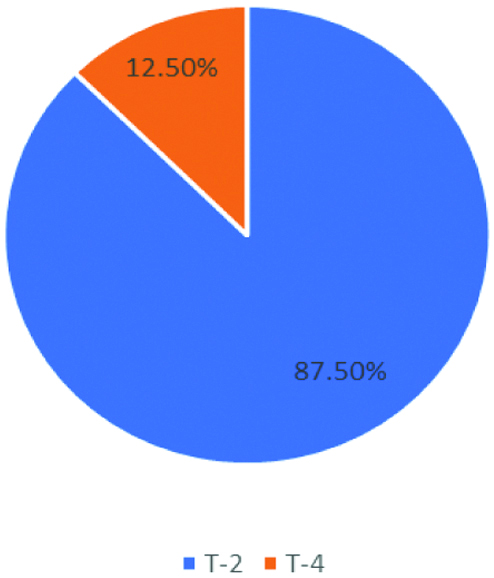
Phage typing was done at Vibrio Phage Reference Lab, NICED, Kolkata by Basu S and Mukherjee S method and New Phage Typing method. According to Basu S and Mukherjee S method, T2 was the most prevalent phage type. Among the isolates, 63 isolates belonged to T-2 phage and 9 isolates were from T-4 phage. None of the strains were untypable [Table/Fig-3] [7].
Distribution of isolates according to Basu S and Mukherjee S classification.
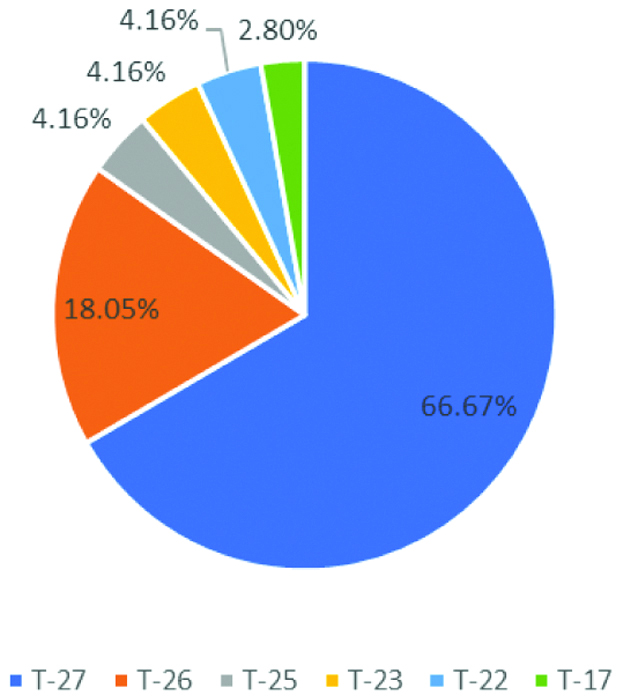
According to New Typing Scheme, 48 isolates belong to T-27 phage. It was the most predominant phage type followed by 13 isolates belonging to T-26, 3 isolates to T-25, T-23, T-22 each and 2 isolates to T-17 [Table/Fig-4].
Distribution of isolates according to the new scheme classification.
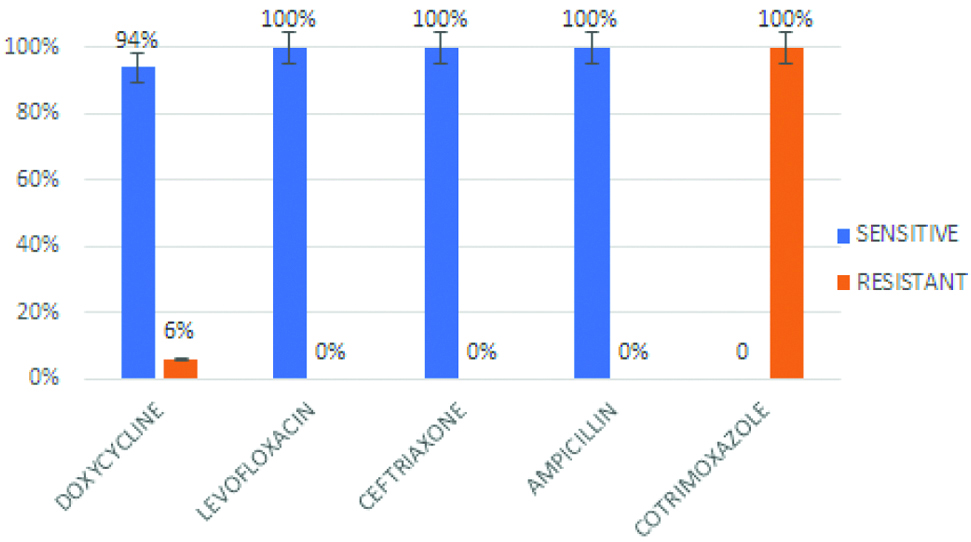
Antibiotic susceptibility was performed by Kirby Bauer disc diffusion method. Resistance to Doxycycline was shown by 6% of isolates. All the isolates were sensitive to Ceftriaxone, Ampicillin and Levofloxacin. Whereas, resistance to Co-trimoxazole was observed in all isolates [Table/Fig-5].
Antibiogram of the Vibrio cholerae O1 isolates.
Discussion
In this study, all the 72 V. cholerae isolates belonged to Serogroup O1, Biotype El Tor and Serotype Ogawa which is in correlation with other studies [12-14]. Kanungo S et al found that V. cholerae O1 belonging to the El Tor was the most common biotype in India and frequency of O139 biotype has been reduced [15]. Phage typing done by old and New method is widely used for epidemiological characterisation of V. cholerae isolates. According to Basu S and Mukherjee S method, the study showed T2 as the predominant Phage type which coincides with studies by Torane V et al., and Turbadkar SD et al., [7,12,16]. However, Sarkar BL et al., reported T4 as predominant phage type [17].
According to New Typing Method, 100% strains are typable. Similar to the study by Turbadkar SD et al., and Sarkar BL et al., with the New Phage Typing method, the study showed T27 as the predominant phage type [16,17]. However, T27 and T25 were reported as predominant strains by Bhowmick TS et al., [18]. Since the study also reported that variation in phage type is associated with resistance to tetracycline, change in antibiotic resistance pattern in V. cholerae isolates can be studied by phage typing [18].
Usually, a seasonal trend in the isolation of V. cholerae is seen. Cases peak in July, taper after September and are negligible after October which closely mimics the seasonal trends in monsoon [12]. However, in present study though the onset of cases coincided with monsoon, they peaked in August and continued till December.
Doxycycline or tetracycline is preferred for the treatment of cholera. This is consistent with the susceptibility pattern over the years [19]. However, the irrational use of these antibiotics could have led to the emergence and spread of the resistant isolates. All the strains showed uniform sensitivity to ampicillin, ceftriaxone and levofloxacin with high level resistance to co-trimoxazole which correlated with other studies [12,14,19]. The phenotypic pattern of the V. cholerae isolates was consistent with other studies. However, a change in the trend of antibiotic susceptibility and seasonal pattern of cases has been observed.
Limitation(s)
The major limitation was that it was conducted only for period of six months. To monitor change in trend of the V. cholerae isolates, continuous monitoring of cases should be carried out which will help to compare the phenotypic trend. Molecular studies are important and should be looked upon for the virulence-associated genes to establish a correlation between biotype and virulence factors.
Conclusion(s)
V. cholerae isolates in this study continue to demonstrate the predominant phenotypic pattern. However, the change in the disease pattern encourage the continued epidemiological and microbiological surveillance. Periodic survey should be carried out to study the trend of phenotypic pattern in the community. The isolates should also be sent to a reference laboratory for routine phage typing.