NTM are mycobacteria which do not cause tuberculsosis. Also, known as atypical Mycobacteria or Mycobacteria other than TB. NTM species are diverse and widespread in the environment, more than 140 NTM species have been identified [1]. In the past, majority of the NTM species were considered as colonisers or environmental contaminants but now increasingly recognised as important pulmonary pathogens in both immunocompromised and immunocompetent population [2]. NTM are now considered as an important cause of lung disease in various parts of the world causing morbidity and mortality [3-5]. NTM often produce infections in the presence of predisposing factors/underlying diseases. The most common being HIV infection, cystic fibrosis, underlying chronic lung disease, previous TB and work in the mining industry [2].
Depending on population and geographic location, NTM infection rates vary widely. Most reports are from developed countries that have low rates of TB [6]. NTM pulmonary disease is often unrecognised or misdiagnosed as MTB and MDR-TB, because of similar clinical presentation in counties with high burden of TB including India. In India due to lack of awareness among clinicians and lack of laboratory facilities to diagnose these infections, its prevalence is largely unknown [6]. Among few reports available in India, NTM isolation rates range from 0.5-8.6% [7]. A recent study from central India reported increasing prevalence from 1.0-3.5% in 2005-2008 [7].
As incidence of antibiotic resistance in pathogens from environment and nosocomial habitats has been increasing, these under documented microorganism needs attention because India is still fighting to overcome the scenario of TB. Most of the clinical studies showed NTM’s to be remarkably resistant or only partially susceptible to the anti-tubercular drugs such as isoniazid, rifampicin, ethambutol and pyrazinamide. Treatment for one species is not effective for others [8]. In India, accurate identification of NTM to the species level and AST is not part of routine laboratory practice, so most of them are treated empirically with anti-TB drugs. The MDR of NTM has now become a globally challenging health issue.
The present study aimed to accurately identify all isolated NTM using different molecular methods and to determine their clinical significance in a TB-endemic region (Telangana) and to know its AST.
Materials and Methods
A prospective study was conducted at Nizam’s Institute of Medical Sciences, Hyderabad, Telangana, India over a period of one year i.e., from June 2017 to May 2018. All the clinical specimens collected from pulmonary and extra-pulmonary TB suspects were included in the study. Institutional Ethics Committee approved the study. The assigned number from ethics committee was SRC/AC4/365, an informed consent was taken from all the patients included in the study. The demographic details like age, sex, occupation, address etc and clinical details such as presenting symptoms, onset of symptoms, previous TB history and co-morbidities were obtained from the subjects. A total of 1085 subjects were included in the study. Sample size was calculated using open Epi web based software using frequency of 8.5% in Indian population.
The clinical specimens were obtained as per the standard protocol [9]. Specimens, which contain normal commensal bacterial flora, were decontaminated with 4% Sodium hydroxide (NaOH) method [9]. All the specimens were subjected to microscopy, culture and geneXpert. The smear microscopy was done using the auramine-rhodamine staining method, culture by BacT/ALERT three-Dimensional (3D) system (BioMerieux, Marcyl’Etoile, France) and Lowenstein-Jensen (LJ) media [10]. The positive flagged bottles were subjected to Zeihl Neelsen (ZN) stain. Samples that failed to show any growth after six weeks of incubation was treated as negative for Mycobacteria. Cultures with positive growth on the BacT/ALERT 3D and presence of Acid-Fast Bacilli (AFB) by ZN stain were tested with a rapid TB antigen assay MPT64 (SD-Bioline Ag MPT64 Rapid TM assay; Standard Diagnostics, Kyonggi-do, Korea) which identifies antigens specific to Mycobacteria Tuberculosis Complex (MTBC). This test was done as per the manufacturer’s instructions [11,12]. The cultures which tested negative for MPT 64 were further characterised by MALDI-TOF and Gene sequencing.
MALDITOF-MS (Matrix Assisted Laser Desorption Ionization Time Of Flight- Mass Spectroscopy)
Loopful of NTM isolates grown on solid medium were taken for each organism. Add 500 μL of 70% ethanol in a pre-labeled 2 mL micro centrifuge vial containing 200 μL of 0.5 mm glass beads and capped securely. A bead beater was used for mechanical disruption of the cells for 5 minutes (or) vortex the tube for 15 minutes and incubated at room temperature for 10 minutes and centrifuged to create a pellet (e.g., 10,000 g for 2 minutes). The pellet was resuspended by adding 10 μL of 70% formic acid to the pellet and repeated aspiration/dispensation was done until the pellet was uniformly dispersed. Add 10 μL of acetonitrile to the vial and mix by repeated aspiration/dispensation, centrifuge and 1 μL of the extract supernatant for each organism was transferred to the designated spots on the target slide. Allow each spot to dry completely. One μL (1 μL) of VITEK MS-CHCA Matrix were transferred to each spot. Allow each spot to completely dry. The slide was run in the MALDI-TOF (BIOMERIEUX VITEC-MS) instrument to obtain the identification [13].
hsp65 Gene Sequencing
Deoxyribonucleic Acid (DNA) from NTM isolates was extracted as described by Van Soolingen D et al., [14]. In short, a loop-full of NTM growth was suspended in 500 μl of 1X Tris-EDTA (TE) buffer (10 mM Tris-HC1, 1 mMEDTA, pH 8.0) and heated at 90°C for 30 minutes in order to lyse the cells. Add 50 μl of lysozyme and incubated for 1 hour at 37°C, add 70 μL of 10% sodium dodecyl sulfate (W/V) and 6 μL of proteinase K (at a 10 mg/mL concentration) the mixture was incubated for 10 minutes at 65°C, followed by phenol-chloroform method used to extract the DNA. The extracted DNA samples were stored at -20°C prior to the next steps. A hsp65 gene sequencing and Polymerase Chain Reaction (PCR) amplification was done to identify the NTM species. hsp65 gene primer sets used were Tb-11 F- ACCAACGATGGTGTCTCCAT and Tb-12R- CTTGTCGAACCGCATACCCT 439 base pairs [15]. PCR reactions were performed using10 mMTris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mMdNTP mix, 0.1 U μl_1 Taq polymerase, 0.5 μM ofeach of the primers, DNA template and nuclease free water.
PCR cycle conditions for amplification of the genes hsp65 were as follows: 94°C for 4 minutes followed by 40 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, and then final extension at 72°C for 10 minutes. Analysis of 3 μL of the PCR products (amplicon) was done by electrophoresis on 1.5% gel agarose. GelRed™ DNA stain was used in staining the gel and the fragments were visualised in the gel documentation system (Gel Doc, ATP Co) under UV light. QIA quick PCR purification kit (QIAGEN, Germany) was used to purify PCR products and analysed by Sanger sequencing (3130 xl-3730 xl genetic analyser, applied biosystem 16/48 capillaries of 50 cm leanth). DNASTAR Laser gene Software (version 7.1) was used to analyse the sequencing results of hsp65 genes. The sequences were compared with similar sequences of the organisms in Gene Bank using the NCBI BLAST online software of the National Center for Biotechnology Information (http://hsp65blast.phsa.ca/blast/blast.html).
Anti-Microbial Susceptibility
All isolates identified by gene sequencing and MALDI-TOF were subjected to antimicrobial susceptibility testing by Broth microdilution as per CLSI guidelines [16]. The following antimicrobials were tested-isoniazid, rifamycin, streptomycin, ethambutol, levofloxacin, moxifloxacin, linezolid, imipenem, co-trimoxazole, clarithromycin, doxycycline and amikacin.
Statistical Analysis
Descriptive statistics included frequencies and prevalence. Continous variables were expressed as median. Categorical variables were expressed as frequencies and percentage. The statistical analysis of the data was done using the microsoft excel and Statistical Package for the Social Sciences (SPSS) version 25.
Results
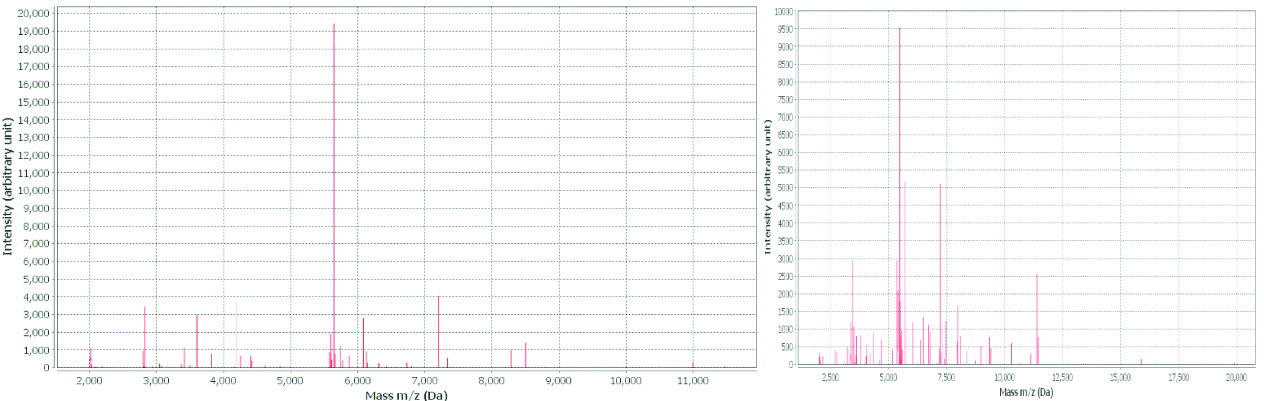
About 1085 samples were processed. A male preponderance was observed (33/55, 60%) while females were 22/55 (40%). The median of age was 40.5 years. The age group 41-50 years was highly affected (41%). Pulmonary samples were predominant 54 out of 55 (98.2%) and one extra pulmonary sample (1.8%). Among the 1085 samples, bronchial wash (27.8%) were maximum followed by followed by tissue, pleural fluid and Cerebrospinal fluid (CSF) [Table/Fig-1]. About 43 patients were associated with co-morbidities. The predominant co-morbidities were bronchiectasis (27.9%) and COPD (25.5%) [Table/Fig-2]. About 15.2% were direct smear positive [Table/Fig-3]. Mycobacterium was detected in 201 cases (18.5%), MTB complex in 146 cases (13.5%) and NTM in 55 (5%) [Table/Fig-4]. Among the 55 NTM isolates, 53 were identified as rapid growers (RGM) 4.8% (M.abscessus) and 2 as slow growers (SGM) 0.18% (M.simiae). MALDI-TOF identified 49 (89%) out of which 48 were M.abscessus and 1- M.simiae [Table/Fig-5].
Sample-wise distribution.
| Sample | Number (n=1085) | Percentage |
|---|
| Bronchial wash | 302 | 27.8 |
| Sputum | 100 | 9.2 |
| Lymph node | 21 | 1.93 |
| Peritoneal fluid | 4 | 0.36 |
| Pleural fluid | 120 | 11 |
| Synovial fluid | 54 | 4.97 |
| Pus | 41 | 3.77 |
| Urine | 84 | 7.74 |
| Tissue | 154 | 14 |
| Gastric aspirate | 6 | 0.55 |
| Ascitic fluid | 32 | 2.94 |
| CSF | 109 | 10 |
| CAPD | 15 | 1.3 |
| BMA | 17 | 1.56 |
| Aspirated fluid | 15 | 1.38 |
| FNAC | 7 | 0.64 |
| Pericardial fluid | 4 | 0.36 |
CSF: Cerebrospinal fluid; CAPD: Continous ambulatory peritoneal dialysis; BMA: Bone marrow aspirate; FNAC: Fine Needle aspiration cytology
Frequency of concomitant co-morbidities in patients (n=43).
| Co-morbidities | Number (n=43) | Percentage |
|---|
| Bronchiectasis | 12 | 27.9 |
| COPD | 11 | 25.5 |
| Previous history of TB | 7 | 16.27 |
| SLE | 5 | 11.6 |
| Malignancy | 4 | 9.3 |
| Sarcoidosis | 2 | 4.65 |
| ILD | 1 | 2.32 |
| Post renal transplant | 1 | 2.32 |
COPD: Chronic obstructive pulmonary disease; SLE: Systemic lupus erythematosis; ILD: Interstitial lung disease; TB: Tuberculosis
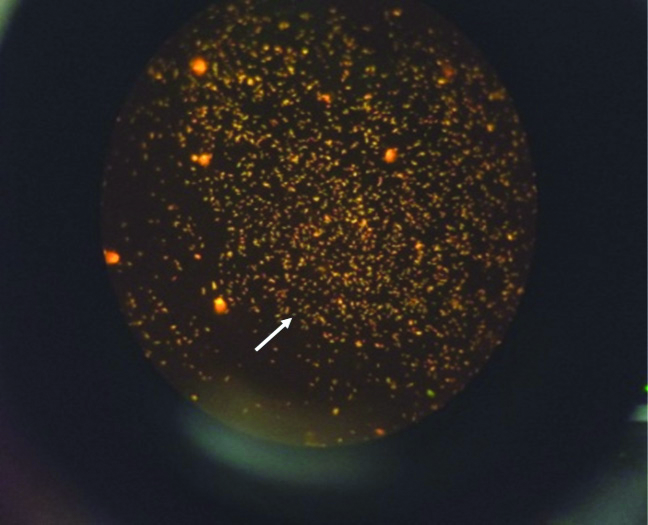
Auramine rhodamine flourescence stain.
The image shows golden yellow fluorescent bacilli
Growth on bact alert, LJ Media and Chocolate agar.

MALDITOF identification of M.abscessus and M.simiae.
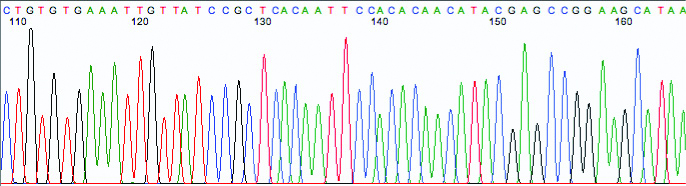
Gene sequencing-partial gene sequencing of hsp65 gene [Table/Fig-6] identified all the 55 (100%) isolates out of which 53 (89%) were M.abscessus and 2 (11%) M.simiae. There was 89% concordance among both the molecular methods [Table/Fig-7]. Subspecies identification of M.abscessus was also done through sequencing where all of them were identified as M.abscessus subspecies abscessus.
Partial hsp65 gene sequencing.
NTM identification by MALDITOF-MS and hsp65 gene sequencing.
| Total no. of isolates | hsp65 gene sequencing identification | MALDITOF-MS identification | % of similarity |
|---|
| 55 | 55 (100%) | 49 (89%) | 89.09% |
| Type strains identified |
| M. abscessus | 53 (96.36) | 48 (97.95) | 90.56% |
| M. simiae | 2 (3.64) | 1 (2.05) | 50% |
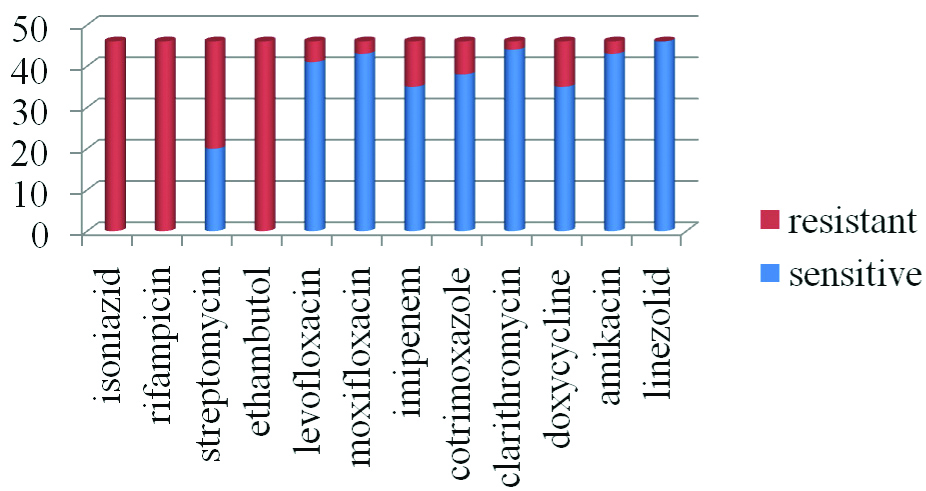
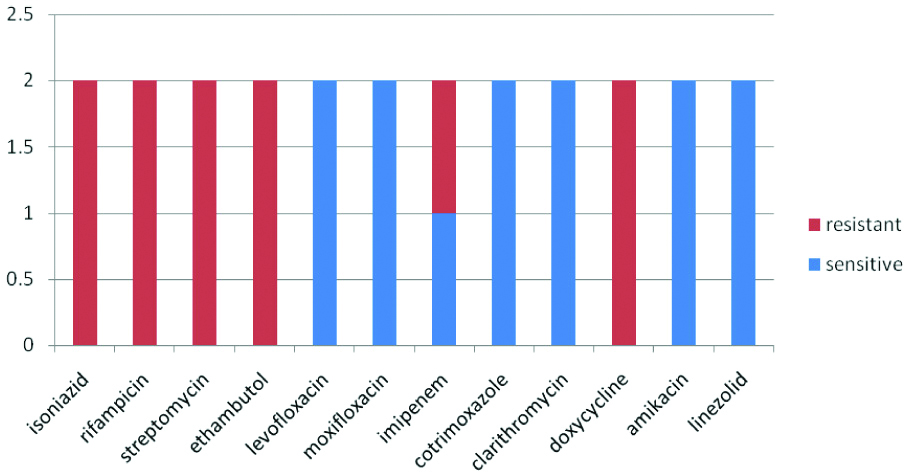
All the identified isolates were subjected to antimicrobial susceptibility testing. M.abscessus showed 100% sensitivity to linezolid followed by clarithromycin, (95.6%), moxifloxacin, (93.4%), amikacin (93.4%). A 100% resistance was observed to rifampicin, isoniazid, ethambutol [Table/Fig-8]. M.simiae showed 100% sensitivity to clarithromycin, amikacin, levofloxacin, moxifloxacin, co-trimoxazole and 100% resistance to the first line drugs [Table/Fig-9].
AST pattern of M.abscessus.
Discussion
Improper identification of NTM can cause clinical and diagnostic dilemmas; just positive microscopy cannot differentiate M.tuberculosis complex from NTM. Management of patients with M.tuberculosis complex and NTM is entirely different therefore prompt isolation, detection and differentiation are necessary for proper and accurate treatment [17,18]. NTM prevalence varies across the globe [Table/Fig-10] [19]. Karak K et al., from Kolkata, have reported a prevalence of 17.4% which was comparatively higher than the other reports [20]. Chakrabarthi A et al., from Chandigarh documented NTM isolation rate of 3.4% from various clinical specimens [21]. Paramasivan CN et al., from Chennai, South India reported 8.6% [22]. Das BK et al., reported isolation of 8.3% NTM from Delhi and Kasauli [23]. The prevalence of NTM was 5% in this study which was closer to the study conducted by Jesudason MV et al., who reported 3.9% [24] [Table/Fig-11].
Worldwide prevalence of NTM [19].
| Regions | Bacteria |
|---|
| USA | MAC, M.kanasasii |
| S.E.America | M.abscessus |
| Europe | MAC |
| N.Europe | M.xenopi |
| UK, Greece | RGM |
| Asia | M.fortuitum |
| E.asia | M.abscessus, M.fortuitum, M.intracellulare |
| Australia | MAC |
| S.australia | RGM (M.abscessus) |
| Africa | M.abscessus, avium, fortuitum |
USA: United States of America; S.E. America: South East America; N.Europe: North Europe; UK: United Kingdom; E.Asia: East Asia; S.Australia: South Australia; MAC: Mycobacterium avium complex; RGM: Rapid grower mycobacteria
Prevalence of NTM-Various studies table [20-22,24-26].
| Study | Prevalence | Specimens | Commonest isolate |
|---|
| Karak K et al., [20] | 17.4% | Sputum | M.fortuitum |
| Chakrabarthi A et al., [21] | 3.4% | Various specimens | M.fortuitum |
| Paramasivan CN et al., [22] | 8.6% | Sputum | M.avium/intracellulare |
| Jesudason MV and Gladstone P [24] | 3.9% | Various specimens | M.abscessus, fortuitum |
| Umrao J et al., [25] | 29% | Various specimens | M.abscessus |
| Jain S et al., [26] | 9.9% | Various specimens | M.kanasasii |
| Current study | 5% | Various specimens | M.abscessus |
In this study, most of the NTM’s were isolated from pulmonary samples, bronchial wash being the predominant sample (95.5%), in Jesudason MV and Gladstone P study recorded largest yield of NTM from pus and biopsy specimens followed by the respiratory specimens [24]. Umrao J et al., study yielded 79.4% from pulmonary sites and 18.2% from extra pulmonary sites, Shenai S et al., study yielded 81% from pulmonary sites [25,27]. The median age group in this study was 40.5 years while Jesudason MV et al., reported 28 years, Umrao J et al., reported 48 years [24,25]. In this study, proportion of males were higher (60%) compared to females which was similar to Umrao J et al., study (60.4%) while Jesudason MV et al., reported 56% males [24,25].
The common co-morbidities in this study were bronchiectasis, COPD and previous history of TB. Jain S et al. also reported main underlying risk factors as COPD (23.1%) and previous TB (30.8%); Shenai S et al., stated COPD 12%, bronchiectasis (9%), previous pulmonary TB (40%) and Huang HL et al., reported previous pulmonary TB (22.1%) and COPD (19.8%) as common co-morbities [26-28]. The direct smear positivity was 15.2% in this study while it was 20% in Shenai S et al., study and 25.6% in the study by Jain S et al., [27,26]. In this study, the culture positivity was 18.5%. Of these positives, 72.6% were identified as MTBC and 27.4% as NTM. Jain S et al., reported 30.7% culture positivity out of which, MTBC constituted (90.1%) and NTM (9.9%) [26]. In Umrao J et al., study, the culture positivity was 19.6%, of which 29.0% were confirmed as NTM and 71.0% MTBC [25]. RGM prevalence was 4.8% while SGM 0.18% in the present study. RGM are a major cause of pulmonary NTM disease in some regions of Asia. This is in contrast with studies of NTM in other parts of the world [2]. The Netherlands study reported only 3% RGM while United States, 5% [29,30]. The high rates in Asia could be due to environmental exposure, isolation frequency and Ethnic factors [28]. Increase in latitude had a direct affect on NTM isolation rates while RGM isolates exhibited a reverse trend [28].
In this study, M.abscessus was the major isolate (95.6%) while Jesudason MV et al., reported M.chelonae-fortuitum as the predominant isolate (87%) and Maurya AK et al., reported M.fortuitum (27.4%) and M.abscessus (14.4%) as the predominant isolates [24,11]. Present study was in agreement with the previous studies carried out in China, Saudi Arabia and Taiwan in which M. abscessus was the predominant NTM species [28]. The MALDI-TOF role in identification of bacteria is already been proved but the challenge is the identification of fungi (yeast and filamentous fungi) and Mycobacteria which are usually difficult with the conventional methods. Molecular methods have shown great improvement in species identification. In this study MALDI-TOF identified 89% of NTM isolates of which M.abscessus is the predominant isolate which is in accordance with Shetye S et al., study 86.67% [31]. In this study, gene sequencing identified all the 55 isolates. MALDI-TOF and gene sequencing identification agreement was 89% which was close to the study conducted by Shetye S et al., 86.67% [31].
The treatment of NTM infections caused by M. abscessus was most difficult [32,33]. In this study 100% sensitivity was observed for linezolid followed by clarithromycin (95.6%), moxifloxacin and amikacin (93.4%). Jesudason MV et al., showed highest susceptibility to amikacin (99.2%) [24]. Ahmed I et al., study RGM isolates showed maximum response to amikacin and clarithromycin, followed by Linezolid, a variable response to Quinolone (45%-ciprofloxacin, 75%- moxifloxacin) and poor sensitivity to Imipenem (20%) [34].
In this study, all the M. abscessus were characterised as subspecies abscessus. Mycobacterium abscessus complex is subdivided into M. abscessus subsp. abscessus, M.abscessus subsp. massiliense, M.abscessus subsp. Bolletii. Inducible macrolide resistance conferred by erythrosomal ribosomal methyl transferase erm (41) gene was observed in M. abscessus and M. bolletii sub species. When M. fortuitum or M. abscessus subsp abscessus isolate is exposed to macrolide, the erm gene activity is induced the organism appear to be susceptible in vitro but will not respond to the macrolide in vivo. This is termed as ‘cryptic resistance which can be ruled out by incubating with macrolide for atleast 14 days [35]. In this study, M.simiae showed 100% resistance to isoniazid, rifampicin, ethambutol, streptomycin and doxycycline and 100% sensitivity to cotrimoxazole, amikacin, clarithromycin, linezolid, moxifloxacin, levofloxacin and 50% resistance to imipenem. Coolen-Allou N et al., study reported maximum susceptibility to amikacin, moxifloxacin, ciprofloxacin and clarithromycin (96%, 92%, 87% and 100% of tested patients, respectively) and resistance to rifampicin, ethambutol and isoniazid were 85%, 89% and 92% respectively [36]. Both slow and rapid growers in this study were susceptible to linezolid, clarithromycin, amikacin and were resistant to first line TB drugs i.e., isoniazid, rifampicin, ethambutol and streptomycin hence AST of NTM’s should be performed as a routine.
Limitation(s)
Study was done for a short duration and follow-up of the patients was not done in the study. Epidemiology and predictors of NTM infections and environmental distribution of NTM need to be studied.
Conclusion(s)
NTM species level identification is done in only specialised laboratories. NTM species shows variation in clinical spectrum and AST. Hence, there is a need to identify NTM isolates to the species level as misdiagnosis can lead to inappropriate treatment. Molecular assay helps in rapid identification which can lead to targeted therapy and can thus combat antimicrobial resistance. The MALDI-TOF also offers quick results at a low cost and is easy to perform, hence it can be considered as an alternate diagnostic tool for identification.
CSF: Cerebrospinal fluid; CAPD: Continous ambulatory peritoneal dialysis; BMA: Bone marrow aspirate; FNAC: Fine Needle aspiration cytology
COPD: Chronic obstructive pulmonary disease; SLE: Systemic lupus erythematosis; ILD: Interstitial lung disease; TB: Tuberculosis
USA: United States of America; S.E. America: South East America; N.Europe: North Europe; UK: United Kingdom; E.Asia: East Asia; S.Australia: South Australia; MAC: Mycobacterium avium complex; RGM: Rapid grower mycobacteria