Infertility is a global phenomenon, about 48.5 million couples are affected by infertility cases [1]. Male factor infertility is reported to account for 20-30% of all the cases of infertility [2]. Chlamydosis, Gonorrhoea, Trichomoniasis and Ureaplasmosis are examples of specific microbial genital infections. Genital infections are mainly caused by Escherichia coli, Staphylococcus spp., Streptococcus spp., Klebsiella spp., and Candida spp. which are known as facultative infections [3,4]. Symptomatic and asymptomatic infertile couples are often evaluated for bacterial infections. These organisms when found in semen are agents of certain forms of morbidity and rarely mortality [5-8].
Infections of the semen may contribute to male infertility by adversely affecting sperm quality and quantity. Bacteria present in the genitals could cause inflammation which may result to oxidative stress, inflammatory scaring and could cause anatomical obstruction and other forms of sperm damage [5-7]. Although the effects of the presence of bacteria on the quality and quantity of semen and sperm cells are unclear, a class of researchers advocates bacterial eradication with antibiotics as a means of improving semen quality among men seeking fertility help with bacterial seminal fluid contamination [8].
Over the years, the pattern of some of these infections, particularly, by the facultative organisms and their resistance profiles are often not reported; apparently due to feelings of non-clinical relevance of such findings [9]. However, there is classical debate and contradictory data on the treatment relevance of these contaminants [10]. Some college of scholars encouraging treatment option are greatly challenged by antibacterial resistance; sometimes expressed as MDR index (i.e., the reciprocal of the total number of resistant antibiotics and the number of antibiotics in the panel tested) [11].
MDR is one of the emerging global public health challenges, as a result of limited antimicrobial treatment options [12-14]. Antimicrobial susceptible bacteria become resistant through a variety of ways such as; chromosomal mutations, Deoxyribonucleic Acid (DNA), uptake through transformation or conjugation and the ability of plasmids to evolve independently [15,16]. In addition, numerous resistance genes from diverse species of bacteria may assemble within a single plasmid and spreads into a wide variety of organisms [14]. MDR of bacteria may be due to functional changes on the cell envelope; which reduces the permeability of the bacterium to antibiotics [17]. Many bacteria resist attack by inactivating drugs through chemical modifications. For example, the hydrolysis of the β-lactam rings of some penicillin by the enzyme penicillinase [18]. The Extended Spectrum Beta Lactamases’ (ESBLs) production by organisms may show cross-resistance to many other antibiotics and thereby limiting therapeutic options [19,20]. Some others may be inactivated by the addition of functional groups. For example, chloramphenicol contains two hydroxyl groups that could be acetylated in a reaction catalysed by the enzyme chloramphenicol acyltransferase with acetyl CoA as the donor [21]. MDR and antibiotic treatment failure has become endemic [15,22]. Some strains of S. aureus that have acquired the mecA gene for Penicillin Binding Protein 2a (PBP2a) are designated as MRSA and are very difficult to eradicate [23]. The enormity of the challenge is increased when the latest licensed antibacterial drug classes-linezolid (1970s) and daptomycin (1980s) were the only alternatives employed for the treatment of MRSA strains, but were barred by their problematic side effects. These left the world with great public health concerns. For instance, S. pneumonia caused about 826, 000 deaths in children aged 1-59 months in 2011 alone, due partly to MDR strain [15]. In a bid to stimulate research and development for newer drugs. WHO published the first ever list of antibiotic resistant “priority pathogens” that pose the greatest threat to human health as: Acinetobacter spp., Pseudomonas spp. and various enterobacteriaceae (including Klebsiella spp. E. coli, Serratia spp. and Proteus spp.) which the Organisation described as critical for Carbapenem resistance and carbapenem resistance/ESBLs production, respectively [22]. Enterococcus faecium, and S. aureus were reported as having high impact on vancomycin resistance and vancomycin/methicillin-resistance, respectively [22].
Globally, it is generally accepted that effective management of pathogens requires empirical knowledge of the antibacterial resistance pattern of local bacterial strains. Antimicrobial resistance surveillance is a public health tool to evaluate the efficacy of antibacterial armamentarium and underscore the need for newer drug discovery. Therefore, in order to provide tools for rational clinical intervention on infections of microbial origin and track the trends of multi-antibacterial resistance, this study was designed to study the current antibiotic susceptibility profile and MDR index of non-specific bacteria from infertile men in Lagos, Nigeria. Present study was the part of author’s previously published study [24].
Materials and Methods
This was a controlled cross-sectional study, involving: (i) selected adult (≥18 years) male patients seeking fertility help; (ii) apparently normal adult male that helped achieve an active pregnancy; and (iii) some repeat male patients from above who were placed and completed antibacterial regimen.
The samples were sourced from the Clinical Science Department, Nigerian Institute of Medical Research, Lagos; the Obstetrics and Gynaecology (O&G) Department, College of Medicine University of Lagos and the Saints Mulumba and David Church infertility clinic, Lawanson, Lagos.
A total of 226 (determined using UNICEF/UNDP/WHO methodology for sample estimation) [25] out of 1,066 infertile men attending fertility clinics screened met the inclusion criteria. One hundred and eight (108) apparently fertile control groups who assisted in verifiable recent pregnancy and 60 patients (from the 226 patients) who were placed on antibiotic treatment and were rescreened after antibiotic treatment regimen were studied, between October, 2009 and December, 2014. The ethical approval was granted by the Institutional Review Board, Nigerian Institute of Medical Research, signed and dated 28th September 2009.
Patients were prequalified through basic physical examination; all patients with other suspected underlying infertility conditions were excluded by a urologist. Those that signed the consent form were included. Patients with poor quality semen were replaced. Those who were unwilling to participate were excluded. For the control group, men of reproductive age who must have assisted to achieve an on-going pregnancy or had sired a child, at least in the last two years were included. All men studied were sexually active, were not on any antibacterial treatment, for at least the past 14 days from the time of inclusion and not on any compelling religious/cultural sexual continence, impediment or obligation.
For the ‘after treatment repeat test group’ all patients with non-specific bacterial infection and antibiotic treatment and repeated visit to the laboratory 14 days after exposure to the antibiotics and completed their regimen as prescribed were included.
Participants were instructed to abstain from sex for at least 3-5 days. They were requested to pass urine first, and then wash hands and the penile surfaces with soap, wash off the soap thoroughly and wipe with a fresh disposable towel before ejaculation of the semen [26,27]. The semen was collected through masturbation using sterile, clean, dry, leak-proof container. Coded identities were employed in handling all specimens to maintain confidentiality.
The specimen was left at room temperature, processed and cultured within one hour of collection [28,29]. Briefly, specimens were cultured onto Nutrient, MacConkey, Chocolate and Blood agar. Samples were incubated in a microaerophilic (5% CO2) and aerobic conditions at 37°C for 18-24 hours [29]. Sample culture were considered positive when the number of colonies was ≥104 CFU/mL for Gram-positive cocci and ≥105 CFU/mL for Gram-negative rods) in the nutrient agar and were identified using the method described by WHO laboratory manual for examination of human semen [26]. Confirmation was by Analytical Profile Index (API) system (Biomerieux, France). Culture for strict anaerobes was not part of this study design. In-vitro antibiotic sensitivity tests were conducted by modified disc diffusion technique, as described by Kirby Bauer, CLSI interpretative criteria [30,31]. Each new batch of agar was tested with a control strain of E. faecalis (ATCC 29212) and Co-trimoxazole disc (1.25 μg + 23.75 μg). The zone of inhibition on standard Mueller-Hinton Agar (MHA) (Oxoid) plate was 20 mm [28,32]. The inoculum was prepared from discrete colony of each isolate to match number 0.5 Mc-Farland standard and the zones sizes of each antimicrobial agent were classified as either: Resistant, Intermediate, and/or Susceptible [30]. The Minimum Inhibitory Concentration (MIC) was conducted using Epsilometer test (E-test) gradient strip (Oxoid). Staphylococcus aureus ATCC 25923 and E. coli ATCC 25922 control strains were used to test the performance of each batch of sensitivity test, while the resistance index was calculated by a method described by Krumperman PH and Olayinka BO et al., respectively [11,33].
Statistical Analysis
Analysis of data was done by Statistical Package for Social Scientists Sciences (SPSS) version 15. Some of the results were expressed in percentage and sperm quantity and quality variables of ‘before and after’ treatment parameters were compared, using a t-test, p-value <0.05 were considered statistically significant at 95% confidence interval.
Results
The socio-demographic characteristics of the population studied is shown in [Table/Fig-1]. Out of the 226 patients, 163/226 (72.1%) had bacterial growth. A total of 229 microbial organisms were isolated. Only 174 (117 Gram positive, 55 Gram negative and 2 yeast-like cells) survived storage in skimmed milk medium with 5% glycerol at -80°C [28].
Socio-demographic factors of the infertile men studied (N=226).
| Patients’ parameters | Number | % |
|---|
| Age (Years) |
| 21-30 | 26 | 11.5 |
| 31-40 | 121 | 53.5 |
| 41-50 | 77 | 34.1 |
| 51 years and above | 2 | 0.9 |
| Education background |
| No formal education | 6 | 2.6 |
| Primary education | 23 | 10.2 |
| Secondary education | 118 | 52.2 |
| Tertiary education | 79 | 35 |
| Duration of infertility |
| ≥1 but <2 years | 47 | 20.8 |
| 2-5 years | 46 | 20.4 |
| ≥6 years | 133 | 58.8 |
| Illness/conditions |
| Alcoholic | 50 | 22.1 |
| Diabetic | 7 | 3.1 |
| Successful surgery | 4 | 1.8 |
| Hypertensive | 8 | 3.5 |
| None | 157 | 69.5 |
| Cancer medication history |
| Yes | 0 | 0 |
| No | 226 | 100 |
| Coffee or caffeinated beverages |
| Yes | 46 | 20.4 |
| No | 180 | 79.6 |
| Occupation |
| Public/civil servant | 72 | 31.9 |
| Trader | 96 | 42.4 |
| Driver/artisan | 40 | 17.7 |
| Others | 18 | 8.0 |
| Childhood infection |
| Chicken pox | 11 | 4.9 |
| Measles | 16 | 7.1 |
| Mumps | 0 | 0 |
| None | 199 | 88 |
| Occupational history |
| Chemical factory | 4 | 1.77 |
| Pesticides factory | 2 | 0.88 |
| None | 220 | 97.35 |
>: Greater than; >: Less than; ≥: Equal to or greater than; Occupation ‘Others’: included persons who claimed to be in school and out of job at that time, poultry farmer and some who live in Lagos but indulge in farming activities in nearby Ogun state; Ethnicity ‘Others’: included persons from South South states, Kwara state, Kogi states and Benue state
Seven persons had multiple microbial growths. Both the organisms that could not survive storage in the refrigerator and multiple growths suspected to be external contaminants were excluded from this analysis.
Only 117/163 (72%) of the patients had antimicrobial prescriptions. A total of 60/163 (36.8%) consented and a repeat visit to the laboratory for cure evaluation was arranged. Of those, 40/60 (66.7%) had only Gram-positive bacterial infection and 20/60 (33.3%) had Gram-negative organisms. Majority, 32/60 (53.3%) had Fluoroquinolone prescribed for them, followed by 17/60 (28.3%) Cephalosporin prescriptions among others. Only 27/60 (45%) persons were treated for 10 days while the majority 33/60 (55%) were treated for up to 14 days in accordance with the clinician’s prescriptions. [Table/Fig-2] shows the occurrence of bacterial isolates.
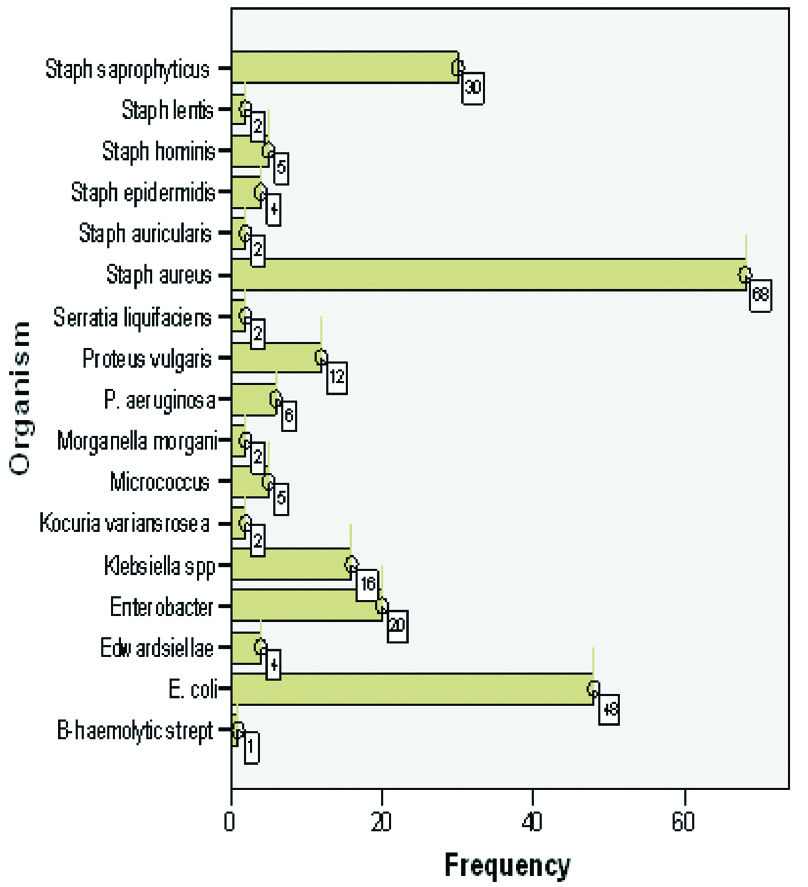
The occurrence of bacterial isolates.
Staphylococcus aureus (Gram positive), E. coli and Enterobacter spp. (Gram negative enterobacteriaceae) were the most prevalent bacterial isolates among the population studied; Both S. aureus and Escherichia coli are established pathogens associated with urogenital inflammation and disease, while Staphylococcus saprophyticus for instance, is seen as nonpathogenic organism (contaminant)
[Table/Fig-3] shows the antibiotic susceptibility/resistance profile of the organisms isolated. The highest sensitive (organisms less resistant) antibiotic from the panel studied was Cefoxitin (90.8%) for Gram-positive cocci e.g., Staphylococcus species and followed by Quinolone (Ciprofloxacin) a broad-spectrum antibiotic that was 54.1% sensitive. The organisms were least susceptible to penicillin with only 1.7% sensitivity (i.e., 98.3% resistance).
Antibiogram range, ranking and frequency percent of antibiotics studied.
| Antibiogram frequency percent |
|---|
| Antibiotics | No zone of inhibition | Zone within resistance range | Total no resistant | % | Zone within intermediate range | Susceptible zone | Total no susceptible | Total % susceptible |
|---|
| No | % | Range mm (equivalent ranking +) | No | % | Range mm (equivalent ranking ++) | No | % | Range mm (Equivalent ranking +++) | No | % |
|---|
| Sulfonamide | 134 | 77.9 | <10 | 10 | 5.8 | 144 | 83.7 | 11-15 | 9 | 5.2 | >16 | 19 | 11 | 28 | 16.3 |
| Chloramphenicol | 130 | 75.6 | <12 | 6 | 3.5 | 136 | 79.1 | 13-17 | 4 | 2.3 | >18 | 32 | 18.6 | 36 | 20.9 |
| Cloxacillin* | 92 | 78.6 | <10 | 13 | 11 | 105 | 89.7 | 11 –12 | 7 | 6.0 | >13 | 5 | 4.3 | 12 | 10.3 |
| Erythromycin | 127 | 73.8 | <13 | 16 | 9.3 | 143 | 83.1 | 14 –22 | 15 | 8.7 | >23 | 14 | 8.1 | 29 | 16.9 |
| Gentamicin (Aminoglycoside) | 91 | 52.9 | <12 | 23 | 13.4 | 114 | 66.3 | 13 -14 | 15 | 8.7 | >15 | 43 | 25 | 40 | 33.7 |
| Amoxicillin/clavunalic acid | 110 | 64 | <13 | 26 | 15.1 | 136 | 79.1 | 14 -17 | 24 | 14 | >18 | 12 | 7 | 36 | 20.9 |
| Streptomycin (Aminoglycoside) | 108 | 62.8 | <11 | 24 | 14 | 132 | 76.7 | 12 -14 | 22 | 12.8 | >15 | 18 | 10.5 | 40 | 23.3 |
| Tetracycline | 138 | 80.2 | <14 | 9 | 5.2 | 147 | 85.5 | 15 -18 | 12 | 7 | >19 | 13 | 7.6 | 25 | 14.5 |
| Amoxicillin (Penicillin) | 167 | 97.1 | <13 | 2 | 1.2 | 169 | 98.3 | 14 -16 | 3 | 1.7 | >17 | 0 | 0 | 3 | 1.7 |
| Nalidixic acid (Quinolone) | 88 | 51.2 | <13 | 62 | 36 | 150 | 87.2 | 14 -18 | 13 | 7.6 | >19 | 9 | 12.8 | 22 | 12.8 |
| Nitrofurantoin# | 28 | 50.9 | <14 | 6 | 10.9 | 34 | 61.8 | 15 -16 | 7 | 12.7 | >17 | 14 | 25.5 | 21 | 38.2 |
| Ceftazidime (Cephalosporin) | 114 | 66.3 | <14 | 28 | 16.3 | 142 | 82.6 | 15 -17 | 25 | 14.5 | >18 | 5 | 2.9 | 30 | 17.4 |
| Ciprofloxacin (Quinolone) | 42 | 24.4 | <15 | 37 | 21.5 | 79 | 45.9 | 16-20 | 49 | 28.5 | >21 | 44 | 25.6 | 93 | 54.1 |
| Cefoxitin (Cephalosporin)$ | 8 | 7.3 | <19 | 2 | 1.8 | 10 | 9.2 | 20 | 22 | 20.2 | >20 | 77 | 70.6 | 99 | 90.8 |
*: Antibiotic unique for Gram-positive organisms; $: Antibiotic unique for 109 Staphylococcus species tested, #: Antibiotic unique for 55 gram-negative organisms; ++ and +++ represent zones of inhibition within susceptible range and outright susceptible range according to CLSI interpretative standard, respectively
[Table/Fig-4] shows antibacterial resistance profile of the specific organisms, within organisms’ total resistance rates of 98.3% AMX, 87.2% NAL and 83.7% COT (for both gram positive and gram negative organisms). The Gram positive specific CXC had resistance rate of 89.7%. Escherichia coli showed 100% resistance rates to AMX, 95.8% ERY, 95.8 % STR, and 91.7% AUG. Tetracycline (TET) had 83.3% and NAL 83.3%. On the other hand, Staphylococcus aureus had resistance rates 97% (AMX), 92.6% (NAL), 91.2% (CXC) among others. Specifically, the study observed that the most prevalent Gram negative organism (E.coli) had 79.2% resistance to COT and 33.3% to CIP. On the other hand, the most prevalent Gram positive organism (S.aureus) had 83.8% and 52.9% resistance to COT and CIP, respectively.
Specific antibacterial resistance profile of the organisms isolated.
| Microorganism | Total no | Antibiotics resistance No (%) |
|---|
| COT | CHL | CXC | ERY | GEN | AUG | STR | TET | AMX | NAL | NIT | CAZ | FOX | CIP |
|---|
| β-haemolytic strept | 1 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) | 1(100) | 0 (0) | NA | 1 (100) | NA | 0 (0) |
| E. coli | 24 | 19 (79.2) | 22 (91.7) | NA | 23 (95.8) | 10 (41.7) | 22 (91.7) | 23 (95.8) | 20 (83.3) | 24 (100) | 20 (83.3) | 14 (58.3) | 16 (66.7) | NA | 8 (33.3) |
| Edwardsiella spp | 2 | 1(50) | 0 (0) | NA | 2 (100) | 1 (50) | 2 (100) | 1 (50) | 1 (50) | 2 (100) | 1 (50) | 1 (50) | 2 (100) | NA | 0 (0) |
| Enterobacter | 10 | 8 (80) | 10 (100) | NA | 10 (100) | 7 (70) | 10 (100) | 10 (100) | 10 (100) | 10 (100) | 9 (90) | 7 (70) | 8 (80) | NA | 7 (70) |
| Klebsieller spp | 8 | 8 (100) | 8 (100) | NA | 7(87.5) | 4 (50) | 7 (87.5) | 6 (75) | 8 (100) | 8 (100) | 7 (87.5) | 3 (37.5) | 7 (87.5) | NA | 6 (75) |
| Kocuria variansosea | 2 | 2 (100) | 2 (100) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | NA | 2 (100) | NA | 2 (100) |
| Micrococcus | 5 | 3 (60) | 5 (100) | 5 (100) | 3 (60) | 5 (100) | 5 (100) | 3 (60) | 5 (100) | 5 (100) | 5 (100) | NA | 5 (100) | NA | 4 (80) |
| Morganella morganii | 1 | 1 (100) | 0 (0) | NA | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 1(100) | 1 (100) | 0 (0) | 1 (100) | NA | 0 (0) |
| P.aeruginosa | 3 | 3 (100) | 3 (100) | NA | 3 (100) | 2(66.7) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 2 (66.7) | 3 (100) | 3 (100) | NA | 0 (0) |
| Proteus vulgaris | 6 | 6 (100) | 6 (100) | NA | 6 (100) | 3 (50) | 6 (100) | 5(83.3) | 5 (100) | 6 (100) | 4 (66.7) | 6 (100) | 6 (100) | NA | 1 (16.7) |
| Serratia liquifaciens | 1 | 0 (0) | 1 (100) | NA | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (50) | NA | 0 (0) |
| Staph. aureus | 68 | 57 (83.8) | 50 (73.5) | 62 (91.2) | 54 (79.4) | 47 (69.1) | 45 (66.2) | 45 (66.2) | 57 (83.8) | 66 (97) | 63 (92.6) | NA | 57 (83.8) | 7 (6.4) | 36 (52.9) |
| Staph. aurecularis | 2 | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 0 (0) | NA | 0 (0) | 0 (0) | 0(0) |
| Staph. epidermidis | 4 | 4 (100) | 4 (100) | 4 (100) | 3 (75) | 4 (100) | 3 (75) | 3 (7) | 4 (100) | 4 (100) | 4 (10) | NA | 3 (75) | 0 (0) | 1(25) |
| Staph. hominis | 5 | 3 (60) | 3 (60) | 5 (100) | 5 (100) | 4 (80) | 4 (80) | 4 (80) | 5 (100) | 5 (100) | 5 (100) | NA | 5 (100) | 0 (0) | 1 (20) |
| Staph. lentis | 2 | 0 (0%) | 0 (0) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 0 (0) | 2 (100) | 2 (100) | NA | 2 (100) | 2 (100) | 2 (100) |
| Staph. saprophyticus | 28 | 26 (92.8) | 20 (71.4) | 22 (78.6) | 22 (78.6) | 22 (78.2) | 23 (82.1) | 21 (75) | 22 (78.6) | 27 (96) | 25 (89.3) | NA | 23 (82.1) | 1 (0.92) | 11 (39.3) |
| Total count | 172 | 144 | 135 | 105 | 143 | 114 | 136 | 132 | 147 | 169 | 150 | 34 | 142 | 10 | 79 |
| % within organism | | 83.7 | 79.1 | 89.7^ | 83.1 | 66.3 | 79.1 | 76.7 | 85.5 | 98.3 | 87.2 | 61.8* | 82.6 | 9.2 | 45.9 |
*: Percentage based on total Gram-negative organisms; NA: Not applicable; ^: Percentage based on Gram-positive organisms; &: percentage based on the 109 Staphylococcus spp; COT: Sulfonamide; CHL: Chloramphenicol; CXC= Cloxacillin; ERY: Erythromycin; GEN: Gentamycin (Aminoglycoside); AUG: Amoxicillin/clavulanic acid; STR: Streptomycin (Aminoglycoside), TET: Tetracycline; AMX: Amoxicillin (penicillin); NAL: Nalidixic acid (Quinolone); NIT: Nitrofurantoin; CAZ: Ceftazidime (cephalosporin); FOX: Cefoxitin (cephalosporin) and CIP: Ciprofloxacin (Fluoroquinolone); GP: Gram positive; GN: Gram negative
[Table/Fig-5] shows Multi Antibacterial Resistance (MAR) index of the bacteria and panel of antibiotics studied. The maximum antibiotic indexes were seen among Enterobacter, Micrococcus and Staphylococcusaureus. The least (0.33) was seen in Serratialiquifaciens.
The Multi Antibacterial Resistance (MAR) index of the sensitivity profile of the bacteria studied.
| Microorganism (n) | Mean multi-drug resistant number | Mean MDR index |
|---|
| β-haemolytic streptococcus (1) | 9 | 0.75 |
| Escherichia coli (24) | 10 | 0.83 |
| Edwardsiella spp. (2) | 8 | 0.67 |
| Enterobacter (10) | 12 | 1 |
| Klebsieller spp. (8) | 11 | 0.92 |
| Kocuria variansosea (2) | 11 | 0.92 |
| Micrococcus (5) | 12 | 1 |
| Morganella morganii (1) | 6 | 0.5 |
| Pseudomonas aeruginosa (3) | 11 | 0.92 |
| Proteus vulgaris (6) | 11 | 0.92 |
| Serratia liquifaciens (1) | 4 | 0.33 |
| Staphylococcus aureus (68) | 13 | 1 |
| Staphylococcus aurecularis (2) | 7 | 0.54 |
| Staphylococcus epidermidis (4) | 11 | 0.85 |
| Staphylococcus hominis (5) | 11 | 0.85 |
| Staphylococcus lentis (2) | 10 | 0.77 |
| Staphylococcus saprophyticus (28) | 11 | 0.85 |
Cure Rate after antibacterial treatment and the ‘after treatment’ quality of the semen were studied. After the treatment, 43 (71.7%) participants were apparently cured (no bacterial presence after 7 days from the last date of antibacterial administration), while 17 (28.3%) were not cured (same species of the previous bacterium isolated).
[Table/Fig-6] shows Sperm quantity and quality variables of ‘before and after’ treatment parameters considering a normal, using a t-test, p-value <0.05 were considered statistically significant at 95% confidence interval. The table demonstrates that despite the high level of resistance in-vitro, administration of antibiotics invivo improved the quality of semen. This underscore the fact that in-vitro resistance may not necessarily translate to invivo resistance, considering the fact that the patients were treated with locally available antibiotics tested.
‘Before and after’ treatment effects: means (SD), range, minimum and maximum parameters of variables from patients exposed to antibacterial treatment.
| Variables | Means±SD | Range | Minimum value | Maximum value | p-value |
|---|
| BT | AT | BT | AT | BT | AT | BT | AT |
|---|
| Sperm concentration in million/mL | 11.548±5.744 | 15.350±6.859 | 18.2 | 28.8 | 0.8 | 1.2 | 19 | 30 | 0.0013* |
| Volume in mL | 2.655±1.1671 | 3.185±0.9165 | 4.5 | 4 | 0.5 | 1.5 | 5 | 5.5 | 0.0066* |
| RPM% | 24.8±16.454 | 33.020±17.234 | 60 | 75 | 0 | 0 | 60 | 75 | 0.0086* |
| SPM% | 19.92±13.662 | 18.00±11.548 | 60 | 50 | 0 | 0 | 60 | 50 | 0.4074 |
| NPM% | 9.28±11.295 | 8.08±8.135 | 50 | 30 | 0 | 0 | 50 | 30 | 0.5056 |
| Immotility % | 45.75±23.449 | 39.90±21.487 | 90 | 92 | 10 | 8 | 100 | 100 | 0.1569 |
| Morphology (>30% abnormal Morphology) | 56.48±25.383 | 51.85±23.784 | 89 | 85 | 10 | 10 | 99 | 95 | 0.3046 |
SD: Standard deviation; RPM: Rapid progressive motility; SPM: Sluggish progressive motility; NPM: Non-progressive motility; BT: Before treatment; AT: After treatment; *<0.01 significance level
Discussion
Generally, the study was on antibacterial sensitivity profile of bacteria from infertile men and has shown that bacterial isolates from infertile semen harbor high MDR strains. It was difficult for people to recollect having had a childhood infection that could affect fertility, as majority had no previous medical records. Only very few described successfully history or scars associated with such previous exposure, mainly through others and not necessarily when they were young.
In this report, S.aureus (29%) and E.coli (10.3 %) were the most prevalent bacterial isolates. This is not different from the trends previously reported in Nigeria by Okon KO et al., and Emokpae MA et al., respectively reported the same as the most prevalent Gram positive and negative bacterial isolates from semen/genitourinary canal [34,35]. In addition, the report from Sri Lanka, bacteria were isolated from 63.6% of the semen samples studied, thus, agreeing with this report of 72.1%. However, the bacteria species were at variance, thus, Streptococcus spp. (36.3%) was the most prevalent, followed by Coliforms, including E.coli (33.3%), Diptheroids (15.1%), Staphylococcus spp. (13.6%), Neisseria spp. (4.5%), and Acinetobacter spp. (1.5%). The prevalence of Staphylococcus spp. was particularly low [36]. The variations may have to do with more stringent biochemical characterisation employed in this study or geographical variance. The notoriety of the duo (Staph. spp. & E.coli) may be associated with the ubiquity of Staphylococcus spp. in anterior nares of certain individuals and human colon respectively. The difference may further underscore the general poor hygiene knowledge, education and practices in most developing communities [37,38].
It is pertinent to highlight that S.aureus and E.coli have been reported to induce sperm damage with two possible putative mechanisms: a direct cytotoxic activity of bacterial toxins and the contact with pili and flagella [39]. Escherichia coli for instance had major resistance trends to β-lactams, Fluoroquinolones and Aminoglycosides; while S.aureus to β-lactams, Fluoroquinolones and Macrolides [40].
From this report, only cefoxitin (90.8%) and quinolone (54%) had sensitivity >50% [Table/Fig-3], the implication is that these antibiotics are still valuable and should be seriously protected from abuse, such that the bacteria will not develop resistance against them. Penicillin had overall least sensitivity of 1.7 % and therefore, apparently of no clinical relevance within the study zone. Among the 14 panel of antibiotics employed; within organism resistance profile for COT, CHL, CXC, ERY, GEN, AUG, STR, TET, AMX, NAL, NIT, CAZ, FOX and CIP were; 83.7%, 79.1%, 89.7%, 83.1%, 66.3%, 79.1%,76.7%, 85.5%, 98.3%, 87.2%, 61.8%, 82.6%, 9.2% and 45.9%, respectively [Table/Fig-4]. All MDR strains studied had MIC as low as 4mg/L to as high as 256 mg/L. Specifically, E.coli showed resistance rates 100% AMX, 95.8% ERY, 95.8% STR, and 91.7% AUG, others are 83.3% TET and 83.3% for NAL. On the other hand, S.aureus had resistance rates 97% for AMX, 92.6% NAL, 91.2% CXC among others. This is particularly worrisome, if we realise that the samples were more like community based isolates, since the patients were merely visiting the clinics for fertility care.
High MAR index of one (1) were recorded among some Gram positive organisms like Staphylococcus, Enterobacter, Micrococcus and Gram negative organisms like Klebsiella spp. (0.92) and E. coli (0.83) studied [Table/Fig-5]. This is particularly significant since the World Health Organisation [22] reported that E.coli species was the most implicated in nonspecific aetiology of UTI globally, causing community and hospital acquired Urinary Tract Infection (UTI), this agrees with the findings of this present study. The WHO further reported the resistance prevalence of >50%; from 5 out of 6 regions of the world, comprising 86/194 member states that provided data for 3rd generation cephalosporin and 92/194-member data for fluoroquinolones for isolates from UTI [15]. Similar profile was experienced in this report; however, the major gap is lack of harmonised control and surveillance pattern in Nigeria when compared with the global best practices.
Specifically, the Western pacific region had E. coli resistance level between 0-70% and Methicillin resistant Staphylococci prevalence of 4-84%. In the South East Asia region, resistance profile of 16-68% for E. coli for 3rd generation cephalosporin and methicillin resistant Staphylococci prevalence of 10-26%; from the Eastern Mediterranean region had 22-63% and methicillin resistant Staphylococci prevalence of 10-53%. Data from Europe were prevalence of 3-82% for E. coli among 3rd generation cephalosporin and fluoroquinolone from 35/36 and 35/35 nations, respectively. Only 31/35 nations returned data for Staphylococci (0.3-60%); same goes for American region with lower resistance rate of 0-48% from 5/7 nations in all for fluoroquinolone and cephalosporin; however, their resistance rate for methicillin resistant Staphylococci (21-90%) was unprecedented among all the regions of the world. Also, 5/7 nations returned data conforming to adequate surveillance system in Europe and America. This is in contrast with what was found among other less developed Asian, Caribbean and African settings [14].
From Asian countries, Marialouis XA and Santhanam A [41], reported phenotypic detection of both ESBLs E. coli and reported higher (84-93%) resistance to nalidixic acid and 100% for amoxicillin/clavulanic acid. Mittal and colleagues equally reported high (95%) and Datta et al., reported lower (88.57%) resistance to amoxicillin/clavulanic acid [42,43]. These reports are not different from the pattern reported in this study and further underscore the need for newer antibiotic discovery.
In Africa, only 13/19 and 14/19 nations returned data to WHO on E. coli for resistance ranging from 2-70% for 3rd generation cephalosporin and fluoroquinolone, respectively [14]. For Staphylococci, 9/15 nation returned data, indicating poor surveillance system in Africa [14]. WHO report on sub-Saharan Africa, Leopold SJ et al., analysed 190 peer reports on resistance in enterobacteriaceae globally and had median prevalence ranged between 31% and 94.2%, whilst median prevalence of resistance to third-generation cephalosporin ranged between 0% and 46.5% [44]. The report concluded high prevalence of resistance to chloramphenicol, trimethoprim/sulfamethoxazole and tetracycline and low prevalence to 3rd generation cephalosporin and fluoroquinolones; this is in agreement with the present report and same with many others [45], comparative resistance trends of E. coli and Staph. aureus from the present study and other studies within Nigeria shown in [Table/Fig-7] [37,38,46-48]. From these studies, it becomes certain the antimicrobial resistance is a global public health menace.
Comparative resistance trends of E. coli and Staph. aureus from the present study and other studies within Nigeria.
| Antibiotic sub-class | Antibiotic | E. coli resistance % | Staph. aureus resistance % |
|---|
| Present study | FMOH (2017) [46] | Nsofor CA and Iroegbu CU [47] | Okonofua FE et al., [38] | Present study | Torimiro N et al., [48] | Shittu AO et al., [37] | Okonofua FE et al., [38] |
|---|
| Penicillin | Amoxicillin | 100 | 48-96 | 94.4 | 100 | 97 | 70 | 75 | 88.2 |
| Amo/Cl, acid | 91.7 | 100 | 74.2 | - | 66.2 | 45 | - | - |
| Cloxacillin | - | 0 -70 | - | - | 91.2 | 85 | - | 16.2 |
| Cephalosporin III | Ceftazidime | 66.7 | 64-91 | - | 0 | 83.8 | 75 | 33 | - |
| Cefoxitin | - | - | 31.5 | - | 6.4 | | - | - |
| Quinolone | Nalidixic acid | 83.3 | - | 38.2 | 0 | 92.6 | 10 | 42 | - |
| Aminoglycoside | Streptomycin | 95.8 | - | 78.7 | 50 | 66.2 | 65 | 75 | - |
| Gentamicin | 41.7 | 85-90 | 24.7 | 50 | 69.1 | 40 | 50 | 14.7 |
| Erythromycin | 95.8 | - | - | - | 79.4 | 40 | - | 11.8 |
| Tetracyclines | Tetracycline | 83.3 | 68-97 | 67.4 | - | 83.3 | 65 | - | 55.9 |
| Fluoroquinolone | Ciprofloxacin | 33.3 | 0 -81 | - | | 52.9 | < 10 | | 29.4 |
| Phenicol | Chloramphenicol | 91.7 | 46-68 | 67.4 | 50 | 73.5 | 25 | 58 | |
| Sulfonamide | Trimethoprim and Sulphamethoxazole | 79.2 | 70 -90 | 85.5 | 50 | 83.8 | 72.1 | 33 | 72.1 |
| Imidazolidines | Nitrofurantoin | 58.3 | - | 70.8 | - | - | - | - | - |
It is necessary to point out that the use of mainly fluoroquinolone and cephalosporin by the patients as prescribed by the clinicians significantly improved some seminal fluid quality, namely: concentration, volume and motility, p-0.0013, 0.0066 and 0.0086 respectively, and this showed that the drugs were efficacious, and that there may be difference between in-vitro and invivo sensitivity presentations.
The resistance rates to the last generation drugs commonly in use to treat serious infections and high MRSA prevalence denote increased risk for patients and this is worrisome. This MDR trend is simply, global. This introduced a need for second-line more toxic drug treatment options with their attendant side effects on the patients. In the geographical area of this study, antibacterial resistance is partly stimulated by selective pressure of sub-lethal dosing, availability of drugs over the counter, the use of antibiotics in animal husbandry and lack of active antimicrobial stewardship teams in the hospitals, among other factors.
It is our candid opinion that antibacterial use and control surveillance group should be made a priority in all African countries where there is none, particularly in Nigeria. Active antimicrobial stewardship teams should be established in every hospital. Drugs should not be dispensed without prescription and the use of antibiotic in animal husbandry should be banned. Finally, Africa may be the custodian of hope to launch the world into the next “magic bullet”, therefore, research and development into discovery of newer antibacterial drugs should be intensified, through good political will and funding.
Limitation(s)
The sample size appears to be not ample, this is an extract of a larger PhD research design involving molecular work. Therefore, cost and stringent inclusion and exclusion criteria affected the sample size.
Conclusion(s)
Although, there is concurrence in the trends of MDR in the study to those seen across the globe, it appears that the resistance prevalence is on the increase because of apparent lack of good policy for surveillance and drug control in Africa in general and Nigeria in particular. Again, going by the recovery rate of the treated group, there is need to exercise strict control on the use of Fluoroquinolone and some Cephalosporin antibacterial armamentarium as reserve drugs in Nigeria in particular and Africa in general.
>: Greater than; >: Less than; ≥: Equal to or greater than; Occupation ‘Others’: included persons who claimed to be in school and out of job at that time, poultry farmer and some who live in Lagos but indulge in farming activities in nearby Ogun state; Ethnicity ‘Others’: included persons from South South states, Kwara state, Kogi states and Benue state
*: Antibiotic unique for Gram-positive organisms; $: Antibiotic unique for 109 Staphylococcus species tested, #: Antibiotic unique for 55 gram-negative organisms; ++ and +++ represent zones of inhibition within susceptible range and outright susceptible range according to CLSI interpretative standard, respectively
*: Percentage based on total Gram-negative organisms; NA: Not applicable; ^: Percentage based on Gram-positive organisms; &: percentage based on the 109 Staphylococcus spp; COT: Sulfonamide; CHL: Chloramphenicol; CXC= Cloxacillin; ERY: Erythromycin; GEN: Gentamycin (Aminoglycoside); AUG: Amoxicillin/clavulanic acid; STR: Streptomycin (Aminoglycoside), TET: Tetracycline; AMX: Amoxicillin (penicillin); NAL: Nalidixic acid (Quinolone); NIT: Nitrofurantoin; CAZ: Ceftazidime (cephalosporin); FOX: Cefoxitin (cephalosporin) and CIP: Ciprofloxacin (Fluoroquinolone); GP: Gram positive; GN: Gram negative
SD: Standard deviation; RPM: Rapid progressive motility; SPM: Sluggish progressive motility; NPM: Non-progressive motility; BT: Before treatment; AT: After treatment; *<0.01 significance level