Introduction
HPV are a family of closely related, epitheliotropic, non-enveloped DNA viruses. Of all the sexually transmitted diseases affecting both men and women, HPV infection tops the chart around the globe [1-4]. The viruses cause a wide range of disease in humans, such as cervical cancer, anogenital cancer, head and neck cancer, skin or genital warts and recurrent respiratory papillomatosis [5-7]. The impact of the disease will rely upon HPV type and site of infection including various host factors.
The HPV has a double-stranded, circular DNA genome of approximately 7900bp and normally encodes two classes of genes: Early (E) genes, E1, E2, E4, E5, E6 and E7, involved in viral replication and cellular transformation and Late (L) genes encoding the L1 and L2: major and minor capsid proteins involved in assembly of viral particles. At present, at least more than 200 HPV genotypes have been currently identified and the whole genome sequence is available in Genbank. These genotypes have been further classified as either high risk or low risk based on their oncogenic potential. The most common low risk subtypes are HPV-6 and 11 whereas the most common high risk subtypes are HPV-16 and 18 [8,9].
Virtually, in all the cases of cervical cancer and 20-50% of the head and neck cancers, HPV plays the central carcinogenic role [10]. The malignancy of the uterine cervix is one of the widely recognised cancers in women according to WHO Information Center on HPV and Cervical Cancer report 2016. The yearly frequency of the disease is practically a large portion of a million and the death pace is roughly half of it. The occurrence pace of cervical cancer in India is roughly 132,000 every year and positions as the second most common malignancy among women in India [11].
Another cancer which is widely attributed to HPV infection is Head and Neck Squamous Cell Carcinoma (HNSCC) especially the ones developed in the oropharynx. HPV-HNSCC has been recognised in the past 10 years as a distinct disease entity and is now responsible for the overwhelming majority of HNSCC in the world. The proportion of HNSCC attributable to HPV is rising substantially with approximately 20-50% of the tumours being HPV positive [9]. Not only is the proportion of HNSCC that are HPV positive rising, but the incidence of HNSCC is also rising. Currently, the incidence of HNSCC (oral cavity, lip and pharynx) is 11.4 per 100,000 populations in India [12].
The foremost important strategy concerning the healthcare system is to put-forth an efficient screening programme for the early detection of viral infection. There are still gaps in knowledge about how HPV drives the cancer progression. Based on the available data, a lot of validated biomarkers for diagnostic and prognostic purposes are used in the clinical management of the subjects suffering from HPV related ailment. With the attributes of efficiently differentiating between various HPV strains with regard to their oncogenic potential, molecular diagnosis has clearly taken over the traditional histopathological and cytological evaluation. The two main purposes served by molecular diagnosis of HPV infection are to conduct a community based screening to identify the population who are at the risk of developing HPV associated malignancy and to follow-up epidemiological studies and vaccination trials [13]. In the view of importance of HPV detection for diagnosis and screening, this review summarises the current molecular tests available that can be harnessed to detect and genotype HPV infection irrespective its site.
Molecular Diagnostics for HPV Detection
Prevention of a disease is the primary goal of any screening programs that continues to be refined as new evidences emerge. Molecular diagnostics is the most effective approach for a cancer control as it aids in the screening of individuals who are prone to that particular cancer.
Molecular assays are the Gold standard for viral DNA detection and necessary in the current scenario as it allows accurate detection and genotyping. Since the link between HPV and cancers is known and numerous large scale studies have proven the point, molecular diagnostics have been introduced into screening algorithms. Propagating HPV in cell culture requires a special model called organotypic raft culture which provides the virus its native epithelial differentiation environment [14,15]. HPV cannot be grown in basic, routine tissue culture and use of serological methods is limited by its decreased accuracy which is not suitable for screening present and past infection. Therefore, HPV detection relies on molecular assays that allow its pinpoint detection and genotyping.
HPV detection and genotyping can be done in a plethora of samples like cervical smears, plasma, saliva, fresh biopsy specimens and FFPE tissue blocks using various molecular techniques. The available molecular assays for HPV detection can be grouped as follows: 1) Signal amplification assays; 2) Nucleic acid amplification assays; and 3) Nucleic acid hybridisation assays.
Signal Amplification Assays
Signal amplification assay which is sometimes called branched DNA assay can be used to facilitate the detection of low abundance molecular targets either in-vivo or in-vitro. Currently, the most reliable signal amplification assays for HPV detection are the FDA approved Hybrid Capture® 2 (HC 2) technology and the Cervista® HPV HR assay kit.
The HC 2 High-Risk HPV DNA Test™ (DNA with Pap™) using Digene Hybrid Capture® 2 (HC 2) assay is an in-vitro nucleic acid hybridisation assay kit [Table/Fig-1] which is capable of amplifying and detecting 13 high risk subtypes of human papilloma virus DNA in cervical specimens, based on the hybridisation of the target HPV-DNA to labelled RNA probes in solution [16,17]. The HPV types that can be detected by using this kit are the high risk HPV types 16,18,31,33,35,39,45,51,52,56,58,59 and 68. The test’s sensitivity is phenomenal, since it can detect DNA of HPV type-16 down to the concentration of 1 pg/mL which relates to 105 viral gene copy numbers. This kit can distinguish between high risk and low risk groups, but is not designed for genotyping single HPV. This is the only drawback of this assay as it does not aid in genotyping of single oncogenic subtype and fails to provide information regarding risk stratification [18].
Representing the workflow of Digene Hybrid capture assay.
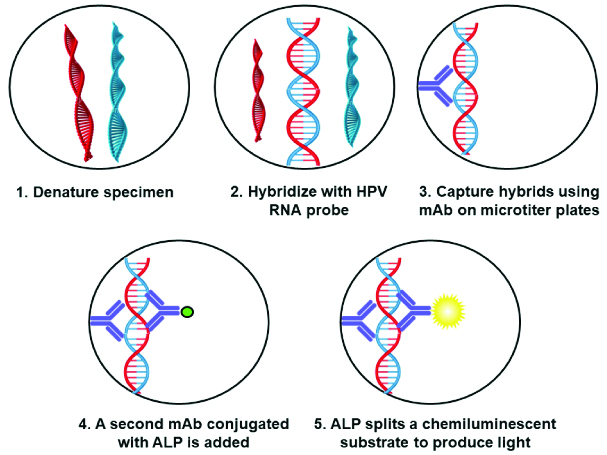
The Cervista® HPV HR test (Hologic, Inc., Marlborough, MA, USA) specifically detects the presence of 14 high risk HPV types comprising of 16,18,31,33,35,39,45,51,52,56,58,59,66 and 68. The assay is based on Invader chemistry, a signal amplification technique for the detection of specific sequences [19,20]. The method represents two isothermal reactions: a) primary reaction on the targeted DNA sequence; and b) secondary reaction that develops a fluorescent signal.
When compared with HC 2, the Cervista® assay demonstrated 100% and 98% sensitivity in the detection of Cervical Intraepithelial Neoplasia (CIN) III and CIN II, respectively [21,22]. Furthermore, this test showed high sensitivity and specificity and a lower false-positive rate in genotyping HPV type-16 and type-18 [23,24].
Nucleic Acid Amplification Assays
Nucleic acid amplification is a crucial process in almost all the molecular biology experiments and has been widely used in forensics, medicine, research and agriculture. Polymerase Chain Reaction (PCR) was the first technique to use nucleic acid amplification and is the preferred method for many application oriented fields as it comes with simple methodology, validated SOP and easier availability of reagents and equipment [25]. Detection of HPV genotypes involves nearly a dozen of nucleic acid amplification assays such as conventional PCR, qPCR, RT PCR, Multiplex PCR, Microarray, PapilloCheck®, Abbott Real time, COBAS 4800 HPV test, CLART HPV 2, INNO-LIPa, Genome sequencing, MCHA, Pre Tect Proofer and APTIMA HPV assay [26].
Generally, HPV detection by PCR can be performed either by type specific primers, designed exclusively to amplify single HPV subtype, or consensus primer pairs, that are designed to amplify a broad spectrum of HPV subtypes. In the recent years, researchers are extensively using the consensus primers such as PGMY09/PGMY11 [27-29] which allows amplification of many types of HPV DNA in a single reaction. These primers are designed to highly conserved L1 region of the HPV genome, which code for major capsid protein [30]. Other consensus primers such as GP5+/GP6+ can be used on the same target to enhance diagnostic sensitivity [31]. General primers in the E1 region have also been described and several other broad-spectrum PCR primers are also reported in the literature [32]. The following are the PCR based nucleic acid amplification assays which are employed in the detection of HPV.
Multiplex PCR is an extension of conventional PCR that involves amplification of multiple targets in a single experiment. This method allows amplification of many target sequences using multiple primer pairs in a reaction mixture [33]. Multiplex Genotyping Kit (Multimetrix, Heidelberg, Germany) is a quantitative and sensitive, high-throughput procedure which involves a PCR-based fluorescent bead array that can detect 24 low and high risks HPV subtypes. The PCR reagents are blended in with the Multiplex HPV Genotyping Kit bead mix. The kit consists of 26 different populations of beads corresponding to 24 HPV probes, 1 β-globin probe and 1 control probe. The resultant PCR products are hybridised onto the probes which are labelled using R-phycoerythrin marked with streptavidin before read on the Luminex analyser [34]. The individual signatures of the beads can recognise the HPV subtypes 6, 11, 16, 18, 26, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73 and 82. Currently, this kit is made available and used for research purposes only. Since it seems to have high sensitivity, it could be applied to large epidemiological studies and has a potential scope in routine diagnostics of HPV [35,36].
Quantitative Real-time PCR (q-RT-PCR/qPCR)
In this post-genomic era, the key to success in viral detection is to have a simple and reliable technology for high-throughput genotyping. More recently, Real-time PCR has brought revolutionary change in the way the clinical laboratories diagnose human pathogens [37,38]. Abbott Real-time High Risk HPV test is a novel method designed to detect 14 (HR-HPV) high-risk human papilloma virus genotypes and aids in distinguishing HPV-16 and HPV-18 from other HR-HPV within a single test [39]. Poljak M et al., evaluated analytical specificity and clinical sensitivity for cervical carcinoma and CIN3 of the Abbott Real-time test in comparison with the Digene Hybrid Capture® 2 Test. The Real-time assay showed excellent analytical specificity and no cross-reactivity with low HPV genotypes that tested positive with Hybrid Capture (HC) 2 and the clinical sensitivity using archived routine cervical specimens was comparable to HC 2 [40].
Microarray Analysis
The microarray-based automated technique permits for parallel detection of multiple DNA samples and is advantageous in gene-expression profiling and mutation analysis [41]. Recently, microarray technology has caught a lot of eye and has been incorporated into the clinical laboratory set-up for HPV detection [42,43]. This technique uses probe amplification, wherein the specific PCR product is hybridised onto a chip and after a washing step, the hybridised signals will be visualised on a DNA chip scanner for HPV DNA detection [44]. PapilloCheck® (Greiner Bio-One GmbH, Germany) is a microarray-based assay, which can detect and identify simultaneously 24 different types of HPV. The assay utilises a multiplex PCR with fluorescent primers designed for the E1 gene of HPV amplifying a 350bp region. Single assay includes 28 probes, each in 5 replicate spots fixed on a DNA chip. Human ADAT1 (adenosine deaminase t RNA specific 1) gene is the internal control and the hybridisation is performed on a microarray chip [45,46]. After hybridisation and repeated washing, the chip is automatically scanned and analysed with the Check scanner™. The HPV types currently detected by PapilloCheck® consists high risk types 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73, 82 and the low risk types 6, 11, 40, 42, 43 and 44 [46]. This assay can be a reliable screening test as it can help in identification of both high risk and low risk HPV and also detect multiple infections. The comparison study between PapilloCheck® with FDA approved Digene HC 2 HPV assay for the detection and identification of 13 High risk HPV types in cervical cancer specimen and anal scrapes shows excellent agreement of 93.8% between two assays [47].
INNO-LiPA (Innogenetics, Ghent, Belgium) is another microarray based assay which detects 14 HPV subtypes. The assay is based on co-amplification of 65bp region of HPV L1 gene and the 270 bp of the human HLA-DPI gene using SPF10 biotinylated primers followed by the hybridisation with specific HPV probes immobilised on a nitrocellulose strip [Table/Fig-2] [48]. Comparative evaluation of Abbott Real-Time and INNO-LiPA for detection of HPV showed complete agreement in detection of 14 common HPV genotypes and partial genotyping of HPV-type 16/18 [49].
Representing the workflow of INNO-LiPA HPV genotyping assay.
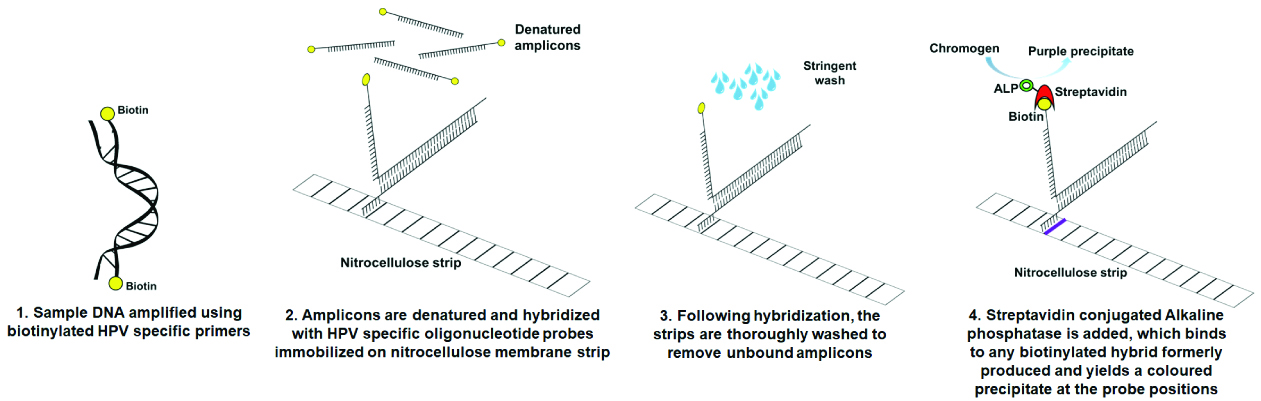
CLART® HPV 2 (Genomica, Madrid, Spain) is based on low density microarray that detects up to 35 HPV genotypes [50,51]. It uses biotinylated primers that amplify a 450 bp fragment within the conserved HPV L1 region to identify 21 high risk types (16, 18, 26, 31, 33, 35, 39, 43, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73, 85 and 89) and 12 low risk types (6, 11, 40, 42, 44, 54, 61, 62, 71, 81, 83 and 84). This assay allows detection of HPV in swabs, cell suspensions and paraffin-wax embedded samples [52].
COBAS® 4800 HPV test is a clinically validated, FDA approved assay that features automated sample preparation combined with real time PCR technology to detect 14 high risk HPV types [53]. The test specifically identifies HPV 16 and 18 while additionally detecting the rest of the high risk types (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) at clinically relevant infection levels [54]. β globin gene is used as internal control for sample adequacy and AmpErase enzyme to protect against PCR cross contamination. HPV primary screening with COBAS® HPV test helps identify women at risk for disease. The landmark registrational ATHENA (Addressing THE Need for Advanced HPV Diagnostics) Study has shown that this test maintains screening efficiency and is better than a Pap (Papanicolaou) test alone as it detects higher grade disease. This test has also proven to be more sensitive than cytology alone for detecting CIN2 and CIN3 [55].
The Linear Array® HPV Genotyping (Roche Molecular Diagnostics, Pleasanton, CA, USA) is based on PCR principle coupled with reverse line blot hybridisation. This assay allows the discrimination of 36 HPV subtypes [56]. The test is directed towards consensus L1 region of the HPV genome to amplify a 450 bp product by using labelled PGMY09/11 primers [57]. To ensure DNA extraction adequacy and PCR efficiency, co-amplification of β-globin gene is used as an internal control. The hybridisation and detection of the amplified product are performed with Auto-LipaTM instrument (Innogenetics, Ghent, Belgium) which can process upto 30 strips simultaneously. Coloured signals are read by the naked eye and the Linear Array® reference guide can be used to interpret the results. When compared with INNO-LiPA assay, this HPV test showed 80.6% agreement between the assays and were highly comparable and reproducible [48].
Microplate Colorimetric Hybridisation Assay (MCHA) (Boehringer Mannheim, Germany) identifies six high risk HPV subtypes 16, 18, 31, 33, 39 and 45 [58]. The assay is based on the amplification by PCR of the 150bp fragment within the L1 region by consensus primers GP5+/GP6+ (General Purpose (GP)), followed by colorimetric hybridisation to six type specific probes on micro well ImmobiliserTM plates [58]. When compared with PapilloCheck®, MCHA showed good agreement for HPV 31, 33 & 45 and also higher sensitivity for identifying HPV 16/18 [59].
HPV-mRNA Detection
Molecular testing of HPV usually involves detection of HPV DNA which indicates the early phase of infection. Research suggests that HPV persists only in a subset of the population which progresses to cancer and hence, it is important to look for secondary markers that have the potential to identify the ongoing transcriptional activity of HPV infection within the population. The early genes E6 and E7 of HPV are mainly responsible for cell transformation and they modulate the activities of cellular proteins such as tumour suppressor proteins p53 and retinoblastoma protein pRb that regulate the cell cycle [60,61]. Therefore, detection of transcriptionally active E6/E7 can be used as a specific marker for diagnosing the precancerous lesions caused by HPV as it serves to be a better predictive marker [62,63]. This could increase the sensitivity and specificity of the screening tests compared with simple detection of HPV-DNA.
Currently, two commercial kits are available to detect the presence of HPV E6/E7 oncogenes: PreTect® Proofer and APTIMA® HPV assay. PreTect® Proofer HPV Assay (NorChip AS, Klok-karstua, Norway) is based on Real-Time multiplex PCR and detects E6/E7 mRNA of 5 high risk HPV subtypes 16, 18, 31, 33 and 45 [64]. The reproducibility of the assay with regard to a positive result was found to be between 96 and 100%. The clinical performance of the kit was analysed with Digene Hybrid Capture® 2 and the PreTect assay showed more specificity in identifying women with CIN2 [65]. APTIMA® HPV assay (Gen-Probe, San Diego, CA, USA) provides better sensitivity than the Proofer assay and is also advantageous as it detects E6/E7 mRNA of 14 high risk HPV subtypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68 [66]. When compared with Hybrid Capture 2 test, APTIMA HPV assay showed similar clinical sensitivity but higher clinical specificity for disease detection which may improve patient management and reduce the cost of care [67,68].
Hybridisation Assays
Hybridisation assays to detect HPV DNA includes Southern blot hybridisation, Dot blot hybridisation and In-situ hybridisation. The first two are old school techniques now, as they come with disadvantages like low sensitivity, time consuming and requiring large amounts of purified DNA [69,70] whereas, in-situ hybridisation allows for precise localisation of specific sequence of nucleic acid within a histologic section. GenPointTM Tyramide Signal amplification system (DAKO, Carpinteria, CA) allows detection as few as 1 or 2 copies of HPV DNA of 13 high risk HPV subtypes (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68). Kelesidis T et al., used it to correlate with the cytological changes and PCR HPV detection, and found that the concordance between In-Situ Hybridization (ISH) and PCR for HPV detection was 78.2% [71]. They also suggested that DNA ISH can detect HPV in cases that may be missed by PCR especially from the formalin fixed paraffin embedded tissues.
As discussed earlier, detection of E6/E7 mRNA transcripts verifies the active viral transcription. RNAscope® HR-HPV assay (Advanced cell diagnostics, Hayward, CA, USA) allows the analysis of RNA on formalin fixed paraffin embedded tissues by utilising oligonucleotide probes for specific for each subtype of high risk HPV E6/E7 mRNA. Dreyer JH et al., evaluated the RNAscope technology by comparing with p16 immunohistochemistry, which is considered surrogate marker for HPV positivity and found a strong correlation between them suggesting the practical evidence of the method [72]. Although commercial kits are marketed for the above methods; the processes are entirely laboratory-based, with access to appropriate reagents and skilled personnel.
Other Emerging Molecular Methods with a Potential to Improvise Current Detection Algorithm
Most of the methodologies discussed above are clinically validated and made commercially available [Table/Fig-3]. Even though these methods show excellent sensitivity and specificity for the detection of HPV infection, there is still need for better molecular methods and biomarkers to identify persistent infections and prevent HPV related cancers. Following are some aspects which are emphasised for the future researchers to consider adding to the existing conventional methods to make the later more sensitive and efficient.
Commercially available assays for HPV detection and genotyping.
| Name of the assay | Assay principle | Nucleic acid target | HPV subtypes detected (HR-High risk, LR-Low risk) |
|---|
| Digene hybrid capture® 2 (hc2) | Signal amplification (microplate chemiluminesence) | DNA | HR-16,18,31,33,35,39,45,51,52,56,58, 59 and 68. |
| Cervista® HPV HR test | Signal amplification (invader chemistry) | DNA | HR-16,18,31,33,35,39,45,51,52,56,58, 59, 66 and 68. |
| Abbott real time high risk HPV test | Nucleic acid amplification (real time PCR) | DNA | Detects 14 HR subtypes and distinguishes between -16 and -18 subtypes. |
| PapilloCheck® | Nucleic acid amplification (multiplex PCR) | DNA | HR- 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73 and 82.LR- 6, 11, 40, 42 and 44. |
| INNO-LiPA | Nucleic acid amplification (multiplex PCR coupled with reverse line blot hybridisation) | DNA | 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68, 69, 70, 73, 74 and 82. |
| CLART® HPV2 | Nucleic acid amplification (Low density microarray) | DNA | HR- 16, 18,26, 31, 33, 35, 39, 43, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73, 85 & 89.LR- 6, 11, 40, 42, 44, 54, 61, 62, 71, 81, 83 and 84. |
| COBAS® 4800 HPV test | Nucleic acid amplification (real time PCR) | DNA | HR- 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68. |
| Linear array® HPV genotyping assay | Nucleic acid amplification (PCR coupled with reverse line blot hybridisation) | DNA | 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72 and 73. |
| PreTect® proofer | Real time multiplex PCR | mRNA | HR- 16, 18, 31, 33 and 45. |
| APTIMA® HPV assay | Real time multiplex PCR | mRNA | HR- 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68. |
Biosensors for HPV Detection
The conventional methodologies used in the detection of HPV genotypes provide robust and reproducible advantages. But, at the same time they involve complex protocols that sometimes are difficult to standardise. Rapid identification of viruses has important implications in healthcare. An ideal instrument would be a hand held biosensor gadget that aids in quantifying the viral particles in the sample and give fast, reliable results [73]. Using these biosensors, researchers can employ virus strains as models to evaluate the interactions between the bio recognition element and the transducer. The device includes immobilisation of antibodies, peptides, aptamers or nucleic acids over the surface of a transducer, which will measure the response signal proportional to the analyte concentration. The signal will be then amplified and processed using computer software. Optical, piezoelectric and electrochemical are the three main types of signal transduction used in biosensors for HPV genotyping [74]. It is essential to evaluate the sensitivity and detection limit of these bio-detection technologies, however, it is also important to consider other parameters such as the specificity, experimental simplicity and cost. Application of biosensors in detection and genotyping of HPV needs further validation in the diagnostics using real samples as blood and other body fluids.
Human Telomerase RNA Component (hTERC) Gene Amplification
It is widely accepted that one of the hallmarks of progressive carcinogenesis is accumulation of chromosomal abnormalities. Gain of function mutation of 3q26 is frequently seen in mucosal originated squamous cell carcinomas. The 3q26 region on the chromosome encodes hTERC gene, whose amplification is believed to be a common event in the progression of cervical cancer through different stages [75]. Liu Y et al., identified amplification of hTERC is important in the development of squamous cell carcinoma of the larynx and suggested it could very well be used as predictive biomarker [76]. The only limitation to this assay is amplification of hTERC is usually identified using techniques such as Fluorescent In-Situ Hybridisation (FISH) and FISH requires highly trained personnel to interpret the staining.
Methylation Profile
DNA methylation plays a vital role in the epigenetic regulation of genes involved in the development of various cancers. Methylation of CpG (C and G trough phosphodiester bond) islands in a gene promoter can essentially lead to silencing of that gene expression. A review by Wentzensen N et al., identified half a dozen of highly heterogeneous genes that promised to be methylation marker candidates for the early detection of cervical cancer [77]. Methylation has been identified in many precancerous stages suggesting it can be used in molecular assay development and over the past years many new high throughput genome wide methylation profiling platforms have been developed.
Conclusion(s)
With the advancement in Molecular Biology, there is scope for rapid growth in the technological improvements to come up with simple, cost-effective and accurate diagnostic tests for diagnosing the viral infection. The molecular assays discussed above show high sensitivity and specificity in detecting and distinguishing high/low risk HPV subtypes and can be used to identify subjects at risk of developing HPV associated cancers in a community based screening program or a clinical setting. However, some of the above described assays are validated only for the use with cervical samples, which could very well applied to obtain knowledge on HPV infection associated with the cancer of the head and neck in a defined population.
[1]. Weinstock H, Berman S, Cates Jr W, Sexually transmitted diseases among American youth: Incidence and prevalence estimates, 2000Perspect Sex Reprod Health 2004 36(1):06-10.10.1363/3600604 [Google Scholar] [CrossRef]
[2]. Koutsky L, Epidemiology of genital human papillomavirus infectionAm J Med 1997 102(5):03-08.10.1016/S0002-9343(97)00177-0 [Google Scholar] [CrossRef]
[3]. Cates Jr W, Estimates of the incidence and prevalence of sexually transmitted diseases in the United States. American Social Health Association PanelJ Sex Transm Dis 1999 26(4 Suppl):S2-7.10.1097/00007435-199904001-0000210227693 [Google Scholar] [CrossRef] [PubMed]
[4]. Hader SL, Smith DK, Moore JS, Holmberg SD, HIV infection in women in the United States: Status at the MillenniumJAMA 2001 285(9):1186-92.10.1001/jama.285.9.118611231749 [Google Scholar] [CrossRef] [PubMed]
[5]. Ljubojevic S, Skerlev M, HPV-associated diseasesClin Dermatol 2014 32(2):227-34.10.1016/j.clindermatol.2013.08.00724559558 [Google Scholar] [CrossRef] [PubMed]
[6]. Crow JM, HPV: The global burdenNature 2012 488(7413):S2-03.10.1038/488S2a22932437 [Google Scholar] [CrossRef] [PubMed]
[7]. Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Global burden of human papillomavirus and related diseasesVaccine 2012 30(Suppl 5):F12-23.10.1016/j.vaccine.2012.07.05523199955 [Google Scholar] [CrossRef] [PubMed]
[8]. Scheurer ME, Tortolero-Luna G, Adler-Storthz K, Human papillomavirus infection: Biology, epidemiology and preventionInt J Gynecol Cancer 2005 15(5):727-46.10.1111/j.1525-1438.2005.00246.x16174218 [Google Scholar] [CrossRef] [PubMed]
[9]. Ramesh PS, Devegowda D, Singh A, Thimmulappa RK, NRF2, p53 and p16: Predictive biomarkers to stratify human papillomavirus associated head and neck cancer patients for de-escalation of cancer therapyCrit Rev Oncol Hematol 2020 1:10288510.1016/j.critrevonc.2020.10288532062315 [Google Scholar] [CrossRef] [PubMed]
[10]. Psyrri A, DiMaio D, Human papillomavirus in cervical and head-and-neck cancerNat Clin Pract Oncol 2008 5(1):24-31.10.1038/ncponc098418097454 [Google Scholar] [CrossRef] [PubMed]
[11]. Update WHOJSR, ICO Information Center on HPV and Cervical Cancer (HPV Information Center): Human papillomavirus and related cancers 2010 [Google Scholar]
[12]. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin D, Piñeros M, Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods 2019 144(8):1941-53.10.1002/ijc.3193730350310 [Google Scholar] [CrossRef] [PubMed]
[13]. Hwang SJ, Shroyer KR, Biomarkers of cervical dysplasia and carcinomaJ Oncol 2012 2012:50728610.1155/2012/50728622131995 [Google Scholar] [CrossRef] [PubMed]
[14]. Doorbar J, Model systems of human papillomavirus-associated diseaseJ Pathol 2016 238(2):166-79.10.1002/path.465626456009 [Google Scholar] [CrossRef] [PubMed]
[15]. Fang L, Meyers C, Budgeon LR, Howett MK, Induction of productive human papillomavirus type 11 life cycle in epithelial cells grown in organotypic raft culturesVirology 2006 347(1):28-35.10.1016/j.virol.2005.10.04316460777 [Google Scholar] [CrossRef] [PubMed]
[16]. Mayrand M-H, Duarte-Franco E, Rodrigues I, Walter SD, Hanley J, Ferenczy A, Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancerN Engl J Med 2007 357(16):1579-88.10.1056/NEJMoa07143017942871 [Google Scholar] [CrossRef] [PubMed]
[17]. Bozzetti MC, Nonnenmacher B, Mielzinska I, Villa L, Lorincz A, Breitenbach V, Comparison between hybrid capture II and polymerase chain reaction results among women at low risk for cervical cancerAnn Epidemiol 2000 10(7):46610.1016/S1047-2797(00)00147-2 [Google Scholar] [CrossRef]
[18]. Stevens MP, Garland SM, Rudland E, Tan J, Quinn MA, Tabrizi SN, Comparison of the Digene Hybrid Capture 2 assay and Roche AMPLICOR and LINEAR ARRAY human papillomavirus (HPV) tests in detecting high-risk HPV genotypes in specimens from women with previous abnormal Pap smear resultsJ Clin Microbiol 2007 45(7):2130-37.10.1128/JCM.02438-0617494721 [Google Scholar] [CrossRef] [PubMed]
[19]. Youens KE, Hosler GA, Washington PJ, Jenevein EP, Murphy KM, Clinical experience with the Cervista HPV HR assay: Correlation of cytology and HPV status from 56,501 specimensJ Mol Diagn 2011 13(2):160-66.10.1016/j.jmoldx.2010.11.01621354050 [Google Scholar] [CrossRef] [PubMed]
[20]. Boers A, Wang R, Slagter-Menkema L, van Hemel BM, Ghyssaert H, van der Zee AG, Clinical validation of the Cervista HPV HR test according to the international guidelines for human papillomavirus test requirements for cervical cancer screeningJ Clin Microbiol 2014 52(12):4391-93.10.1128/JCM.02716-1425297324 [Google Scholar] [CrossRef] [PubMed]
[21]. Boers A, Slagter-Menkema L, van Hemel BM, Belinson JL, Ruitenbeek T, Buikema HJ, Comparing the Cervista HPV HR test and Hybrid Capture 2 assay in a Dutch screening population: Improved specificity of the Cervista HPV HR test by changing the cut-offPloS one 2014 9(7):e10193010.1371/journal.pone.010193025051098 [Google Scholar] [CrossRef] [PubMed]
[22]. Johnson LR, Starkey CR, Palmer J, Taylor J, Stout S, Holt S, A comparison of two methods to determine the presence of high-risk HPV cervical infectionsAm J Clin Pathol 2008 130(3):401-08.10.1309/4DXEAFG2JXYF34N3 [Google Scholar] [CrossRef]
[23]. Einstein MH, Martens MG, Garcia FA, Ferris DG, Mitchell AL, Day SP, Clinical validation of the Cervista® HPV HR and 16/18 genotyping tests for use in women with ASC-US cytologyGynecol Oncol 2010 118(2):116-22.10.1016/j.ygyno.2010.04.01320488510 [Google Scholar] [CrossRef] [PubMed]
[24]. Bartholomew DA, Luff RD, Quigley NB, Curtis M, Olson MC, Analytical performance of Cervista® HPV 16/18 genotyping test for cervical cytology samplesJ Clin Virol 2011 51(1):38-43.10.1016/j.jcv.2011.01.01621376660 [Google Scholar] [CrossRef] [PubMed]
[25]. Abreu AL, Souza RP, Gimenes F, Consolaro ME, A review of methods for detect human Papillomavirus infectionVirol J 2012 9:26210.1186/1743-422X-9-26223131123 [Google Scholar] [CrossRef] [PubMed]
[26]. Zaravinos A, Mammas IN, Sourvinos G, Spandidos DA, Molecular detection methods of human papillomavirus (HPV)Int J Biol Markers 2009 24(4):215-22.10.1177/17246008090240040120108214 [Google Scholar] [CrossRef] [PubMed]
[27]. Remmerbach TW, Brinckmann UG, Hemprich A, Chekol M, Kühndel K, Liebert UG, PCR detection of human papillomavirus of the mucosa: comparison between MY09/11 and GP5+/6+ primer setsJ Clin Virol 2004 30(4):302-08.10.1016/j.jcv.2003.12.01115163418 [Google Scholar] [CrossRef] [PubMed]
[28]. Haws ALF, He Q, Rady PL, Zhang L, Grady J, Hughes TK, Nested PCR with the PGMY09/11 and GP5+/6+ primer sets improves detection of HPV DNA in cervical samplesJ Virol Methods 2004 122(1):87-93.10.1016/j.jviromet.2004.08.00715488625 [Google Scholar] [CrossRef] [PubMed]
[29]. Bandhary SK, Shetty V, Saldanha M, Gatti P, Devegowda D, SR Pushkal, Shetty AK, Detection of human papilloma virus and risk factors among patients with head and neck squamous cell carcinoma attending a tertiary referral centre in South IndiaAsian Pac J Cancer Prev 2018 19(5):1325 [Google Scholar]
[30]. Lungu O, Wright TC, Silverstein S, Typing of human papillomaviruses by polymerase chain reaction amplification with L1 consensus primers and RFLP analysisMol Cell Probe 1992 6(2):145-52.10.1016/0890-8508(92)90059-7 [Google Scholar] [CrossRef]
[31]. Ramesh PS, Devegowda D, Naik PR, Doddamani PA, Nataraj SM, Evaluating the feasibility of nested PCR as a screening tool to detect HPV infection in saliva of Oral Squamous Cell Carcinoma subjectsJ Clin Diagn Res 2018 12(7):BC22-25.10.7860/JCDR/2018/34880.11806 [Google Scholar] [CrossRef]
[32]. Tieben LM, terSchegget J, Minnaar R, Bavinck JNB, Berkhout RJ, Vermeer BJ, Detection of cutaneous and genital HPV types in clinical samples by PCR using consensus primersJ Virol Methods 1993 42(2-3):265-79.10.1016/0166-0934(93)90038-S [Google Scholar] [CrossRef]
[33]. Edwards MC, Gibbs RA, Multiplex PCR: Advantages, development and applicationsGenome Res 1994 3(4):S65-75.10.1101/gr.3.4.S658173510 [Google Scholar] [CrossRef] [PubMed]
[34]. Ozaki S, Kato K, Abe Y, Hara H, Kubota H, Kubushiro K, Analytical performance of newly developed multiplex human papillomavirus genotyping assay using Luminex xMAP™ technology (Mebgen™ HPV Kit)J Virol Methods 2014 204:73-80.10.1016/j.jviromet.2014.04.01024768623 [Google Scholar] [CrossRef] [PubMed]
[35]. Schmitt M, Bravo I, Snijders PJ, Gissmann L, Pawlita M, Waterboer T, Bead-based multiplex genotyping of human papillomavirusesJ Clin Microbiol 2006 44(2):504-12.10.1128/JCM.44.2.504-512.200616455905 [Google Scholar] [CrossRef] [PubMed]
[36]. Schmitt M, Dondog B, Waterboer T, Pawlita M, Tommasino M, Gheit T, Abundance of multiple high-risk human papillomavirus (HPV) infections found in cervical cells analyzed by use of an ultrasensitive HPV genotyping assayJ Clin Microbiol 2010 48(1):143-49.10.1128/JCM.00991-0919864475 [Google Scholar] [CrossRef] [PubMed]
[37]. Kaltenboeck B, Wang C, Advances in real-time PCR: Application to clinical laboratory diagnosticsAdv Clin Chem 2005 40(4):219-59.10.1016/S0065-2423(05)40006-2 [Google Scholar] [CrossRef]
[38]. Deepak S, Kottapalli K, Rakwal R, Oros G, Rangappa K, Iwahashi H, Real-time PCR: Revolutionizing detection and expression analysis of genesCurr Genomics 2007 8(4):234-51.10.2174/13892020778138696018645596 [Google Scholar] [CrossRef] [PubMed]
[39]. Huang S, Tang N, Mak W-B, Erickson B, Salituro J, Li Y, Principles and analytical performance of Abbott Real Time High Risk HPV testJ Clin Virol 2009 45(suppl. 1):S13-S7.10.1016/S1386-6532(09)70003-4 [Google Scholar] [CrossRef]
[40]. Poljak M, Oštrbenk A, Seme K, Učakar V, Hillemanns P, Bokal EV, Comparison of clinical and analytical performance of the Abbott realtime high risk HPV test to the performance of hybrid capture 2 in population-based cervical cancer screeningJ Clin Microbiol 2011 49(5):1721-29.10.1128/JCM.00012-1121430098 [Google Scholar] [CrossRef] [PubMed]
[41]. van Hal NL, Vorst O, van Houwelingen AM, Kok EJ, Peijnenburg A, Aharoni A, The application of DNA microarrays in gene expression analysisJ Biotechnol 2000 78(3):271-80.10.1016/S0168-1656(00)00204-2 [Google Scholar] [CrossRef]
[42]. Hwang TS, Jeong JK, Park M, Han HS, Choi HK, Park TS, Detection and typing of HPV genotypes in various cervical lesions by HPV oligonucleotide microarrayGynecol Oncol 2003 90(1):51-56.10.1016/S0090-8258(03)00201-4 [Google Scholar] [CrossRef]
[43]. An HJ, Cho NH, Lee SY, Kim IH, Lee C, Kim SJ, Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with the HPV DNA chip microarray methodCancer 2003 97(7):1672-80.10.1002/cncr.1123512655524 [Google Scholar] [CrossRef] [PubMed]
[44]. Rays M, Chen Y, Su YA, Use of a cDNA microarray to analyse gene expression patterns in human cancerNat Genet 1996 14(4):457-60.10.1038/ng1296-4578944026 [Google Scholar] [CrossRef] [PubMed]
[45]. Dalstein V, Merlin S, Bali C, Saunier M, Dachez R, Ronsin C, Analytical evaluation of the PapilloCheck test, a new commercial DNA chip for detection and genotyping of human papillomavirusJ Virol Methods 2009 156(1):77-83.10.1016/j.jviromet.2008.11.00219041893 [Google Scholar] [CrossRef] [PubMed]
[46]. DNA-Chip H. PapilloCheck® HPV-Screening. 2006 [Google Scholar]
[47]. Didelot MN, Boulle N, Damay A, Costes V, Segondy M, Comparison of the PapilloCheck® assay with the digene HC2 HPV DNA assay for the detection of 13 highrisk human papillomaviruses in cervical and anal scrapesJ Med Virol 2011 83(8):1377-82.10.1002/jmv.2214821678441 [Google Scholar] [CrossRef] [PubMed]
[48]. van Hamont D, van Ham MA, Bakkers JM, Massuger LF, Melchers WJ, Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the roche linear array HPV genotyping testJ Clin Microbiol 2006 44(9):3122-29.10.1128/JCM.00517-0616954236 [Google Scholar] [CrossRef] [PubMed]
[49]. Kocjan BJ, Seme K, Poljak M, Comparison of the Abbott RealTime High Risk HPV test and INNO-LiPA HPV Genotyping Extra test for the detection of human papillomaviruses in formalin-fixed, paraffin-embedded cervical cancer specimensJ Virol Methods 2011 175(1):117-19.10.1016/j.jviromet.2011.04.00621513740 [Google Scholar] [CrossRef] [PubMed]
[50]. Bonde J, Rebolj M, Ejegod DM, Preisler S, Lynge E, Rygaard C, HPV prevalence and genotype distribution in a population-based split-sample study of well-screened women using CLART HPV2 Human Papillomavirus genotype microarray systemBMC Infect Dis 2014 14(1):41310.1186/1471-2334-14-41325064473 [Google Scholar] [CrossRef] [PubMed]
[51]. Poljak M, Cuzick J, Kocjan BJ, Iftner T, Dillner J, Arbyn M, Nucleic acid tests for the detection of alpha human papillomavirusesVaccine 2012 30:F100-F6.10.1016/j.vaccine.2012.04.10523199952 [Google Scholar] [CrossRef] [PubMed]
[52]. Pista A, Verdasca N, Oliveira A, Clinical performance of the CLART human papillomavirus 2 assay compared with the hybrid capture 2 testJ Med Virol 2011 83(2):272-76.10.1002/jmv.2195221181922 [Google Scholar] [CrossRef] [PubMed]
[53]. Heideman D, Hesselink AT, Berkhof J, van Kemenade F, Melchers WJG, Daalmeijer NF, Clinical validation of the cobas® 4800 HPV Test for cervical screening purposesJ Clin Microbiol 2011 49(11):3983-85.10.1128/JCM.05552-1121880968 [Google Scholar] [CrossRef] [PubMed]
[54]. Abraham J, Stenger M, Cobas HPV test for first-line screening for cervical cancerJ Community Support Oncol 2014 12(5):156-57.10.12788/jcso.003924971425 [Google Scholar] [CrossRef] [PubMed]
[55]. Wright TC, Stoler MH, Behrens CM, Apple R, Derion T, Wright TL, The ATHENA human papillomavirus study: Design, methods, and baseline resultsAm J Obstet Gynecol 2012 206(1):46.e1-.e11.10.1016/j.ajog.2011.07.02421944226 [Google Scholar] [CrossRef] [PubMed]
[56]. Dobec M, Bannwart F, Kilgus S, Kaeppeli F, Cassinotti P, Human papillomavirus infection among women with cytological abnormalities in Switzerland investigated by an automated linear array genotyping testJ Med Virol 2011 83(8):1370-76.10.1002/jmv.2212621678440 [Google Scholar] [CrossRef] [PubMed]
[57]. Giuliani L, Coletti A, Syrjänen K, Favalli C, Ciotti M, Comparison of DNA sequencing and Roche Linear Array® in human papillomavirus (HPV) genotypingAnticancer Res 2006 26(5B):3939-41. [Google Scholar]
[58]. Barcellos RB, de Matos Almeida SE, Sperhacke RD, Verza M, Rosso F, de Medeiros RM, Evaluation of a novel microplate colorimetric hybridisation genotyping assay for human papillomavirusJ Virol Methods 2011 177(1):38-43.10.1016/j.jviromet.2011.06.01021807028 [Google Scholar] [CrossRef] [PubMed]
[59]. Keegan H, Pilkington L, McInerney J, Jeney C, Benczik M, Cleary S, Human papillomavirus detection and genotyping, by HC2, full-spectrum HPV and molecular beacon real-time HPV assay in an Irish colposcopy clinicJ Virol Methods 2014 201:93-100.10.1016/j.jviromet.2014.02.00224583109 [Google Scholar] [CrossRef] [PubMed]
[60]. Narisawa-Saito M, Kiyono T, Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: Roles of E6 and E7 proteinsCancer Sci 2007 98(10):1505-11.10.1111/j.1349-7006.2007.00546.x17645777 [Google Scholar] [CrossRef] [PubMed]
[61]. Zur Hausen H, Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesisJ Natl Cancer Inst 2000 92(9):690-98.10.1093/jnci/92.9.69010793105 [Google Scholar] [CrossRef] [PubMed]
[62]. Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS, High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridisation assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinomaAm J Sur Pathol 2011 35(9):1343-50.10.1097/PAS.0b013e318220e59d21836494 [Google Scholar] [CrossRef] [PubMed]
[63]. Castle PE, Dockter J, Giachetti C, Garcia FA, McCormick MK, Mitchell AL, A cross-sectional study of a prototype carcinogenic human papillomavirus E6/E7 messenger RNA assay for detection of cervical precancer and cancerClin Cancer Res 2007 13(9):2599-605.10.1158/1078-0432.CCR-06-288117473189 [Google Scholar] [CrossRef] [PubMed]
[64]. Molden T, Kraus I, Skomedal H, Nordstrøm T, Karlsen F, PreTect™ HPV-Proofer: Real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomavirusesJ Virol Methods 2007 142(1):204-12.10.1016/j.jviromet.2007.01.03617379322 [Google Scholar] [CrossRef] [PubMed]
[65]. Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of the Hybrid Capture 2 test for identification of women at risk of cervical cancerJ Clin Microbiol 2010 48(8):2779-85.10.1128/JCM.00382-1020573862 [Google Scholar] [CrossRef] [PubMed]
[66]. Dockter J, Schroder A, Hill C, Guzenski L, Monsonego J, Giachetti C, Clinical performance of the APTIMA® HPV Assay for the detection of high-risk HPV and high-grade cervical lesionsJ Clin Virol 2009 45(Suppl. 1):S55-S61.10.1016/S1386-6532(09)70009-5 [Google Scholar] [CrossRef]
[67]. Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, Aptima HPV E6/E7 mRNA test is as sensitive as Hybrid Capture 2 Assay but more specific at detecting cervical precancer and cancerJ Clin Microbiol 2011 49(2):557-64.10.1128/JCM.02147-1021147950 [Google Scholar] [CrossRef] [PubMed]
[68]. Arbyn M, Roelens J, Cuschieri K, Cuzick J, Szarewski A, Ratnam S, The APTIMA HPV assay versus the hybrid capture 2 test in triage of women with ASC-US or LSIL cervical cytology: A meta-analysis of the diagnostic accuracyInt J Cancer 2013 132(1):101-08.10.1002/ijc.2763622610699 [Google Scholar] [CrossRef] [PubMed]
[69]. Kuypers JM, Critchlow C, Gravitt PE, Vernon D, Sayer J, Manos M, Comparison of dot filter hybridisation, Southern transfer hybridisation and polymerase chain reaction amplification for diagnosis of anal human papillomavirus infectionJ Clin Microbiol 1993 31(4):1003-06.10.1128/JCM.31.4.1003-1006.19938385147 [Google Scholar] [CrossRef] [PubMed]
[70]. Villa LL, Denny L, Methods for detection of HPV infection and its clinical utilityInt J Gynaecol Obstet 2006 94(Suppl. 1):S71-S80.10.1016/S0020-7292(07)60013-7 [Google Scholar] [CrossRef]
[71]. Kelesidis T, Aish L, Steller MA, Aish IS, Shen J, Foukas P, Human papillomavirus (HPV) detection using in situ hybridisation in histologic samples: Correlations with cytologic changes and polymerase chain reaction HPV detectionAm J Clin Pathol 2011 136(1):119-27.10.1309/AJCP03HUQYZMWATP21685039 [Google Scholar] [CrossRef] [PubMed]
[72]. Dreyer JH, Hauck F, Oliveira-Silva M, Barros MH, Niedobitek G, Detection of HPV infection in head and neck squamous cell carcinoma: A practical proposalVirchows Arch 2013 462(4):381-89.10.1007/s00428-013-1393-523503925 [Google Scholar] [CrossRef] [PubMed]
[73]. Frías IA, Avelino KY, Silva RR, Andrade CA, Oliveira MD, Trends in biosensors for HPV: Identification and diagnosisJournal of Sensors 2015 2015:91364010.1155/2015/913640 [Google Scholar] [CrossRef]
[74]. Caygill RL, Blair GE, Millner PA, A review on viral biosensors to detect human pathogensAnal Chim Acta 2010 681(1):08-15.10.1016/j.aca.2010.09.03821035597 [Google Scholar] [CrossRef] [PubMed]
[75]. Sui W, Ou M, Dai Y, Chen J, Lan H, Yan Q, Gain of the human telomerase RNA gene TERC at 3q26 is strongly associated with cervical intraepithelial neoplasia and carcinomaInt J Gynaecol Obstet 2009 19(8):1303-06.10.1111/IGC.0b013e3181b62ea520009881 [Google Scholar] [CrossRef] [PubMed]
[76]. Liu Y, Dong Xl, Tian C, Liu H-g, Human telomerase RNA component (hTERC) gene amplification detected by FISH in precancerous lesions and carcinoma of the larynxDiagn Pathol 2012 7(1):3410.1186/1746-1596-7-3422463766 [Google Scholar] [CrossRef] [PubMed]
[77]. Wentzensen N, Sherman ME, Schiffman M, Wang SS, Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-scienceGynecol Oncol 2009 112(2):293-99.10.1016/j.ygyno.2008.10.01219054549 [Google Scholar] [CrossRef] [PubMed]