Polymyxins (polymyxin B and colistin) are the last resort antimicrobial agents for the treatment of serious bacterial infections such as pneumonia, sepsis, meningitis, pyelonephritis, bacterial gastro-enteritis, osteomyelitis etc., caused by MDR Enterobacteriaceae, in particular Carbapenem-Resistant Enterobacteriaceae (CRE). The main mechanisms associated with acquired polymyxin resistance are associated to mutations within the regulatory systems that modify the Lipopolysaccharide (LPS) of the bacterial outer membrane, leading to a decrease in electrostatic interactions between the polymyxins and therefore the bacterial membrane, triggering the resistance [1,2]. Recently, Liu YY et al., described the primary mobile polymyxin resistance mechanism, mediated by the gene Mobile Colistin Resistance-1 (mcr-1) located in an exceedingly plasmid. The decrease in the LPS affinity for polymyxins is due to the mcr-1 protein of phosphoethanolamine transferase family which promotes the addition of phosphoethanolamine to lipid A (lipopolysaccharide component) [3]. The mcr-1 gene causes an excellent concern about its dissemination among polymyxin susceptible Enterobacteriaceae. Regardless of the resistance mechanism, the distribution of polymyxin resistant strains limits the therapeutic options, resulting in high mortality rates. The usual routine microbiology susceptibility tests (disk diffusion and gradient diffusion test) are not reliable to judge the susceptibility to polymyxins. The very fact is that the sole method considered accurate for the detection of resistance to polymyxins {the Broth Microdilution (BMD)} is laborious and expensive, therefore the development of fast and reliable method is needed [4-6]. Recently, Nordmann P et al., developed the Rapid Polymyxins NP Test. This test detects the bacterial growth on glucose in the presence of a defined concentration of polymyxin B or colistin. Growth of bacteria lead to glucose metabolism, which will change the pH and acidify the medium detected by visible colour. The result is obtained within four hours, although majority of the resistant isolates showed positive results in around two hours [6]. The aim of this study was to assess the performance of the Rapid Polymyxins NP test in the detection of polymyxin resistance among clinical isolates of E.coli and K.pneumoniae and compare the resistance pattern detected by VITEK-2.
Materials and Methods
This cross-sectional study was conducted in the Department of Microbiology of Govind Ballabh Pant Institute of Postgraduate Medical Education and Research (GIPMER), New Delhi, India from April 2019 to August 2019. Informed consent was not taken as no procedure was done on the patient and hence it was not required. Institutional Ethical Committee (IEC) number was obtained-2019/10/07-03.
Various non-duplicate clinical samples such as blood, urine, sputum, pus and endotracheal aspirates received in the laboratory were inoculated on blood agar and MacConkey agar plates and incubated at 37°C for 18-24 hour and processed according to standard bacteriological procedures [7,8]. On the basis of the growth on blood agar and MacConkey agar, isolates were thereafter processed in VITEK-2 systems, for identification and antimicrobial susceptibility testing. E.coli and K.pneumoniae isolated from above samples were further processed by Rapid Polymyxin NP test. Samples having pure growth of single organism namely E.coli and K.pneumoniae were included in the study and samples with mixed growth were not included in the study.
The antibiotic susceptibility test was performed by using VITEK-2 compact systems (bioMerieux, France). Interpretive breakpoints for Colistin (MIC ≤2 μg/mL is susceptible, and MIC ≥4 μg/mL is resistant) were used as per CLSI guidelines [9].
Results of identification and antimicrobial susceptibility test of organism were obtained after 16-18 hours.
Rapid Polymyxin NP Test
Test method: The Rapid Polymyxins NP test was performed according to Nordmann P et al., [6]. The antibiotic solutions (colistin at 0.2 mg/mL) were mixed with the Rapid Polymyxin NP solution (2.5% of Mueller Hinton broth-cation adjusted powder, 0.005% of phenol red indicator and 1% of D (+)-glucose) into sterile glass tubes in a proportion of 1:40. Given that, the final concentration containing colistin into tubes was 5 μg/mL [Table/Fig-1]. The bacterial inoculum was standardised to obtain a 3.0-3.5 McFarland optical density (≈109 CFU/mL) in sterile solution of sodium chloride (NaCl) 0.85%. The test was performed in a 96-well polystyrene plate and for each isolate, 50μL of the bacterial suspension was inoculated in two wells: 1st well only 150 μL of the Rapid Polymyxin NP solution (free of antibiotics), 2nd well 150 μL of the Rapid Polymyxin NP solution with colistin. The final concentration of bacteria was adjusted to 108 CFU/mL in each of the wells, and therefore the final concentration of colistin sulfate was 3.75 μg/mL. The plate was incubated for up to four hour at 35°C±2°C and the first reading was made at 15 minutes and then every hour.
Representative results of the rapid polymyxin NP [Nordmaan/Poirel] test. Noninoculated wells are shown as controls (first column I). The rapid polymyxin NP test was performed with a reference colistin-susceptible isolate as negative control (second column II) and with a reference colistin-resistant isolate as positive control (third column III) in a reaction medium without (upper row) and with (lower row) colistin. The tested isolate grew in the presence (and absence) of colistin (wells B IV and A IV, B V and A V, B VI and A VI respectively) and was therefore reported to be colistin-resistant.
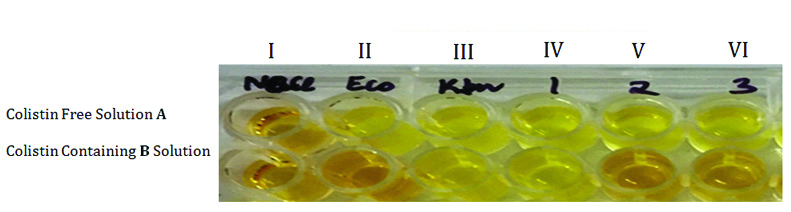
Interpretation: The test was considered positive (polymyxin resistance) when the isolate grew in presence of colistin, changing the color of the Rapid Polymyxin NP solution containing antibiotic from orange to yellow, i.e., the same color of the well containing only NP solution. However, the test was considered negative (polymyxin susceptible) when the isolate did not grow in the presence of antibiotics and the color of the solution remained orange [6].
Quality control: A known carbapenemase-producing Klebsiella pneumoniae (confirmed by PCR) was used as positive control, and E. coli ATCC 25922 was used as negative control.
Invalid test: The test was considered invalid if there occurs a spontaneous change in colour of the microtitre plate within 10 minutes of putting the test [6].
Statistical Analysis
Kappa analysis was done to evaluate the percentage agreement between Vitek-2 and Rapid Polymyxin NP tests.
Results
Three hundred and ten isolates were analysed in the present study, out of which 75% (232) strains were E.coli and 78 were K. pneumoniae. Most strains of E.coli 127 (54.74%) and K.pneumoniae 27 (34.61%) were isolated from pus and body fluids [Table/Fig-2].
Sample wise distribution of isolates.
| Sample | E.coli (232) | K.pneumoniae (78) |
|---|
| Pus/ Drain fluid | 127 | 27 |
| Blood | 43 | 26 |
| Body fluids | 10 | 5 |
| Endotracheal aspirate | 52 | 20 |
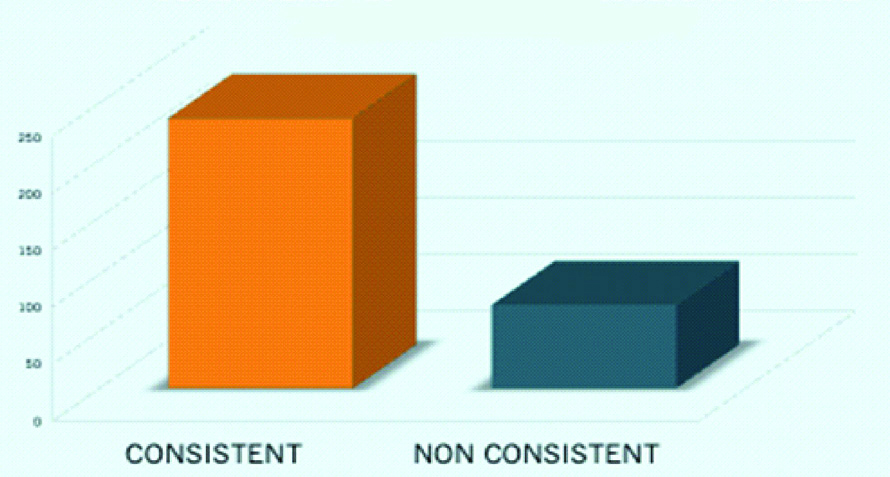
Above strains were tested for colistin resistance by Vitek-2 and Rapid Polymyxin NP test. The results of both methods were consistent in (237/310) 76.45% cases and non-consistent in (73/310) 23.55% cases [Table/Fig-3]. Kappa analysis revealed that the strength of agreement between the two test was considered moderate (kappa=0.412, confidence interval: from 0.312 to 0.513).
Consistency between the results.
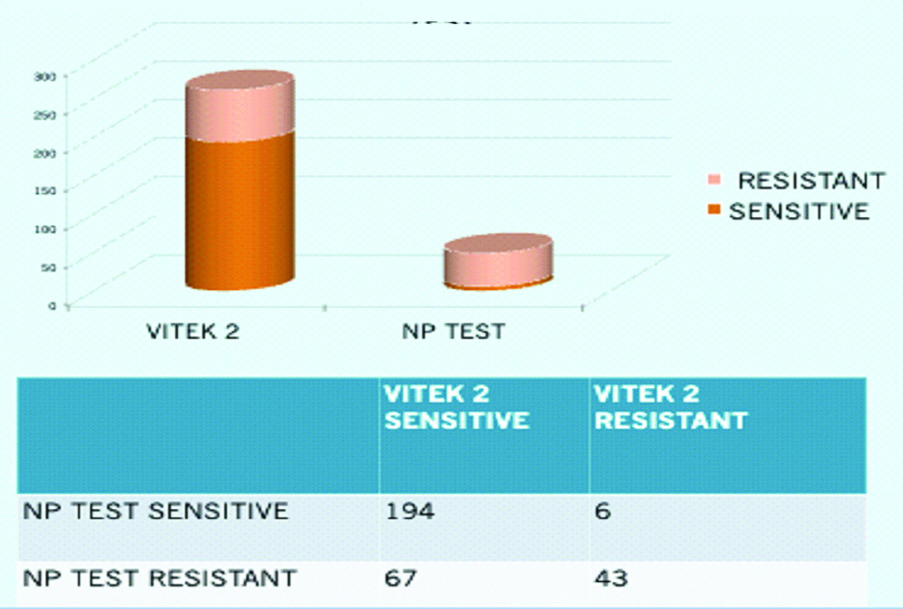
A total of 194 (62.58%) isolates were sensitive by both VITEK-2 and Rapid Polymyxin NP Test. Forty-three (13.87%) isolates were resistant by VITEK-2 and Rapid Polymyxin NP Test. Six (1.93%) isolates which were resistant by VITEK-2, later on came to be sensitive by rapid polymyxin NP test. Sixty-seven (21.61%) isolates which were sensitive by VITEK-2 were found to be resistant by Rapid Polymyxin NP test [Table/Fig-4]. The sensitivity and the specificity of Rapid Polymyxin NP test as compared to VITEK-2 was 74.32% and 12.24%, respectively.
Comparison between Vitek-2 and Polymixin NP.
Discussion
Emergence of infections caused by MDR gram-negative bacteria is on a rising trend worldwide. To control the emergence of drug resistance, the irrational use and misuse of antibiotics should be controlled. Rapid detection and reporting of drug resistance pattern in clinical isolates is one of the way which helps in the selection of appropriate antibiotics for treatment. The increasing resistance to many antibiotics limits a lot of therapeutic options and has led to an increase in the use of intravenous colistin in tertiary care setting, therefore this study was taken up to evaluate Rapid Polymyxin NP test for rapid detection of polymixin resistance.
Out of 310 isolates, 232 were E. coli and 78 were K. pneumoniae. The result of both methods was consistent in 76.45% cases with moderate strength of agreement. Sixty seven (21.61%) isolates which were sensitive to Colistin by VITEK-2 were found to be resistant by Rapid Polymyxin NP test. Hence, Vitek-2 should not be used to report sensitivity to Colistin.
VITEK-2 is a semi-automated system that uses reagent cards containing dehydrated antibiotics and other reagents in a 64-well format. It combines rapid identification and antibiotic susceptibility testing based on an extrapolated growth algorithm.
Similar findings have been reported by Tan TY and Ng SY; they recommended that VITEK-2 colistin susceptibility test to be an unreliable method [10]. However studies done by Lo-Ten-Foe JR et al., stated that colistin susceptibility was well performed by VITEK-2. The VITEK-2 colistin susceptibility test is also considered to be a reliable tool to work out susceptibility to colistin in isolates that don’t exhibit resistant subpopulations [11]. Despite the actual fact, the VITEK-2 is straight forward to use susceptibility testing method within the diagnostic microbiology laboratory, care should be taken within the interpretation of the results for genera during which hetero resistance has been described (Enterobacter and Acinetobacter). For these genera, an alternate testing method capable of detecting resistant subpopulations such as E-test and agar dilution test should be used [11].
Rapid Polymyxin NP test is a much faster technique in comparison to the reference method BMD. In comparison, BMD is laborious, time consuming and relatively expensive. In general, for 300 isolates the time taken for the test to become positive was two hours. Although 10 isolates showed positive in one hour and only one isolate needed three hours to show positive results. The sensitivity and specificity of the Rapid Polymyxins NP test was 98%. This test was easy to perform and considerably fast as shown in other studies too [4,6]. In studies done by Jayol A et al., they read the colour change of the wells every hour and found that final results were obtained two hours after incubation when the tray was incubated at 35±2°C under an ambient atmosphere [4]. However, positive results (frank color change) were obtained as early as 1 hour after incubation for Klebsiella spp. and E. coli isolates. Isolates which are resistant, gave positive results after 1 hour of incubation when incubated trays at 35±2°C under 5% CO2 [6]. These study confirmed that the test provides results much faster than the BMD (12-18 hours). It can contribute to the rapid identification of polymyxin-resistant isolates. Additionally in this study, Rapid polymyxin NP test showed excellent sensitivity and specificity, which was in similarity with the results found in the study of Nordmann P et al (sensitivity 99.3% and specificity 95.4%) [12]. However, in this study sensitivity and specificity couldn’t be calculated as BMD was not performed.
In a study conducted in Greece, the efficiency of Rapid Polymyxin NP test was tested against K.pneumoniae, including 98 colistin-resistant and 33 colistin-susceptible isolates and the performance of this test was compared with automated systems like BD Phoenix, VITEK-2. The Rapid Polymyxin NP had accurately detected 97 out of 98 colistin-resistant isolates except one isolate of K. pneumoniae harboring a wild-type mgrB gene, yielding a sensitivity of 99%. The opposite methods gave more false-negative results with colistin-resistant strains. Also, BD Phoenix, VITEK-2 and also the gradient E-test have missed colistin-resistant strains. In comparison, Rapid Polymyxin NP test gave false positive results with only six isolates, yielding the specificity of 82%. Despite the fact that Rapid Polymyxin exhibited lower specificity than other methods, the rapidness of this method can’t be ignored. Thus, these findings indicate that the Rapid Polymyxin NP test will be an initial weapon for the detection of colistin-resistant isolates in near future [13].
Several studies have shown that, Rapid Polymyxin NP test showed excellent performance for the detection of resistant isolates and susceptible Enterobacteriaceae for colistin, when compared to the reference method [Table/Fig-5] [4,14,15]. Additionally, the time needed to get the result reduced from approximately 24 hours (BMD test) to around two hours (Rapid Polymyxins NP test), besides, being less laborious. [Table/Fig-6] shows the advantages and disadvantages of Rapid Polymyxin NP test and VITEK-2. The rapid identification of polymyxin-resistant isolates may contribute for more precise therapy choices, likewise because the rapid implementation of contact precaution measures, preventing the event of outbreaks with MDR organisms.
Studies showing excellent Rapid Polymyxin NP test [4,14,15].
| Study | Year | Sensitivity | Specificity |
|---|
| Jayol A et al., [4] | 2016 | 99.3% | 95.4% |
| Jayol A et al., [14] | 2018 | 98.1% | 94.9% |
| Malli E et al., [15] | 2019 | 98.1% | 94.9% |
Advantages and disadvantages of rapid NP and VITEK-2.
| Rapid NP test | VITEK-2 system |
|---|
| Advantages | 1. Rapid turn around time=4 hrs2. Cost-effective | 1. Less labour intensive2. MIC given3. No observer bias |
| Disadvantages | 1. Observer bias2. Labour intensive3. Expertise setting required4. Detects only resistance | 1. Expensive2. Turnaround time=16-18 h |
Limitation(s)
Limitation of this study was that the author did not compare Rapid polymyxin NP test with BMD. However, we plan to compare Rapid polymyxin NP test with BMD in the future research for E.coli and K.pneumoniae isolated from clinical samples.
Conclusion(s)
Rapid polymyxin NP test when compared to VITEK-2 is simple, easy to perform, sensitive, specific and cost-effective method. Thus, it can be used in laboratories for screening of polymyxin resistance in carbapenemase-producing Gram-negative bacteria as recommended by CLSI. Also, the automated VITEK-2 System showed a variable performance to detect the susceptibility to colistin hence, its results should be reconfirmed by Rapid polymyxin NP test or BMD test.
Declaration: This article is an original work and was previously presented in the form of a poster at the 11th Annual Conference of IAMM Delhi chapter held at India habitat center, New Delhi on 2 November 2019.