Enterococci, Gram positive, facultative anaerobic cocci, have evolved over the past few decades, from being intestinal commensals of man and animals to becoming important nosocomial pathogens associated with significant morbidity and mortality [1,2]. They cause a wide range of infections like Urinary Tract Infections (UTI), Surgical Site Infections (SSI), bacteremia, intra-abdominal and intra-pelvic abscess, and occasionally, meningitis and pneumonia.
Since its first report in 1988, the emergence of Vancomycin Resistant Enterococci (VRE) as one of the leading cause of nosocomial infection globally is of particular concern, as it has limited the therapeutic options available for the clinicians [2,7-10]. Furthermore, Enterococci act as reservoirs of these antibiotic resistance genes and it tends to transfer these genes to other bacteria including Methicillin-Resistant Staphylococcus aureus (MRSA) [11]. Over a 15 year period there was a 20-fold increase in VRE associated with nosocomial infections reported to CDC’s National Nosocomial Infections Surveillance [12]. In North America and Europe, VRE rates are about 30%, with most isolates being E. faecium (>90%) [12,13]. Compared to the western data, the VRE isolation rate in India ranges from 0.89%-10% of Enterococcal isolates [7,11,14,15].
Even though there is ample data from other parts of the country on the prevalence of Vancomycin resistance and HLGR in Enterococci, there is a paucity of data from this state and especially this geographical area. The knowledge of the prevalence of such drug resistant strains can help the clinicians in prescribing the appropriate drug combinations; eliminating the therapeutic challenge of these organisms due to their ease of acquiring and transference of drug resistance.
Hence, the present study was conducted in at tertiary care teaching hospital at Thrissur, Kerala, to identify Enterococcal species causing various infections in this particular geographical area and their antibiotic resistance pattern with respect to HLGR and VRE.
Materials and Methods
A hospital based cross-sectional study was done in the Department of Microbiology, Government Medical College, Thrissur, Kerala, India. The study was done with the permission of Institutional research Committee and Institutional ethics review Board of the institution. Based on the previous laboratory records of Government medical College, Thrissur, a sample size of 60 was calculated, but as 75 Enterococci were isolated during the study period, all the isolates were included. All clinically relevant Enterococci isolated from various clinical specimens from both inpatients and outpatients, received in the Department of Microbiology during the period of study were included in the study after taking informed consent from the patients. The isolated strains were considered clinically significant and included in the study, when obtained in pure culture or in significant numbers as part of mixed cultures; and the isolates from stool samples were excluded.
A total of 75 consecutive culture isolates of Enterococci from January 2017 to December 2017 were collected and speciation and antimicrobial susceptibility were done simultaneously during the same period. The isolates were obtained from clinical samples like blood, urine, pus and other body fluids like pleural fluid, peritoneal fluid, CSF (Cerebrospinal fluid). Samples were inoculated on to Blood and MacConkey agar and incubated at 37°C for 24-48 hours aerobically. On the 2nd and 7th day of incubation, blood samples in Brain heart infusion broth were subcultured on to Blood and MacConkey agar [Table/Fig-1,2]. Preliminary identification of Enterococci was done by colony morphology, Gram stain, and catalase test [16]. Further speciation was done by Bile Aesculin test, Pyrrolidonyl Arylamidase (PYR) test, Growth in presence of 6.5% Sodium chloride, Growth at 10°C and 60°C and Heat resistance test [16].
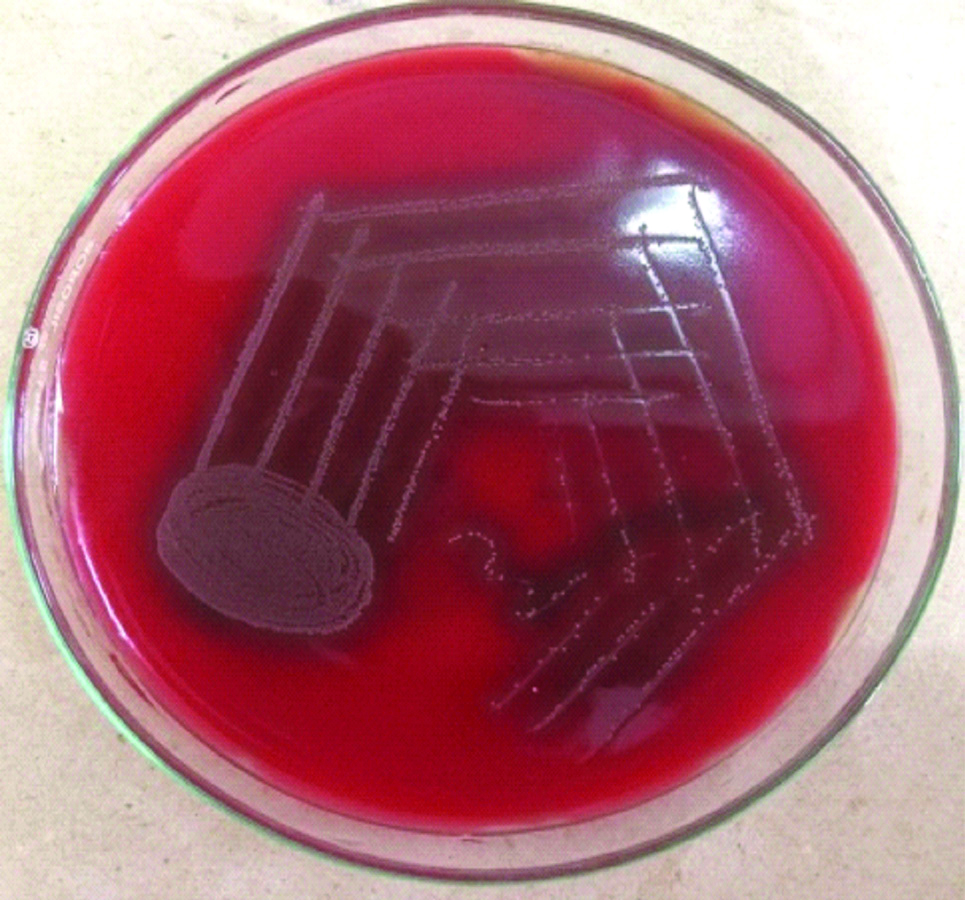
Growth E. faecalis on blood agar.
Alpha haemolytic colonies on blood agar
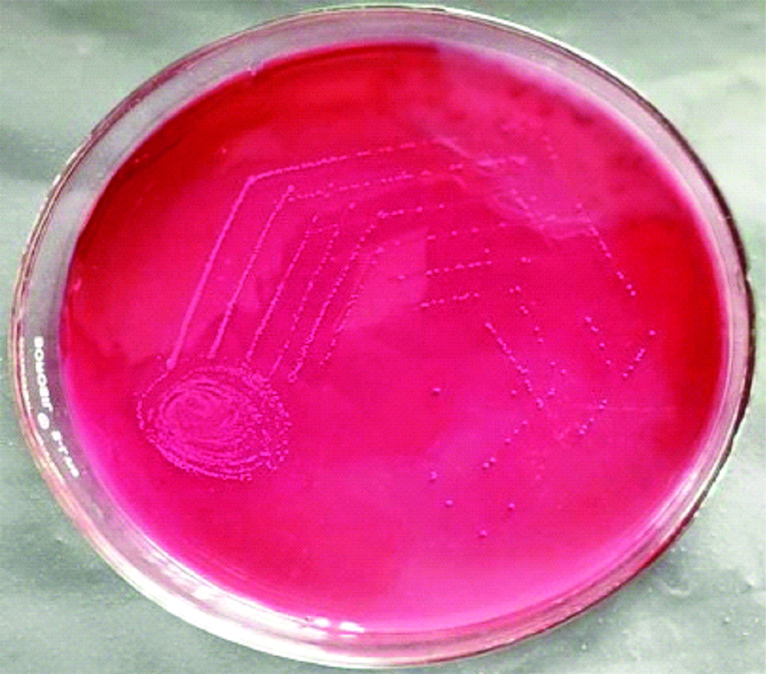
Growth E. faecalis on mac conkey agar.
Magenta coloured colonies on Mac conkey agar
The species of Enterococcus can be separated into five groups based on conventional biochemical tests like acid formation from mannitol and sorbose and hydrolysis of arginine [4]. Species determination was done by:
i) Fermentation of sugars-1% glucose, sucrose, maltose, lactose, mannitol, arabinose and raffinose.
ii) Arginine hydrolysis by Moller decarboxylase test.
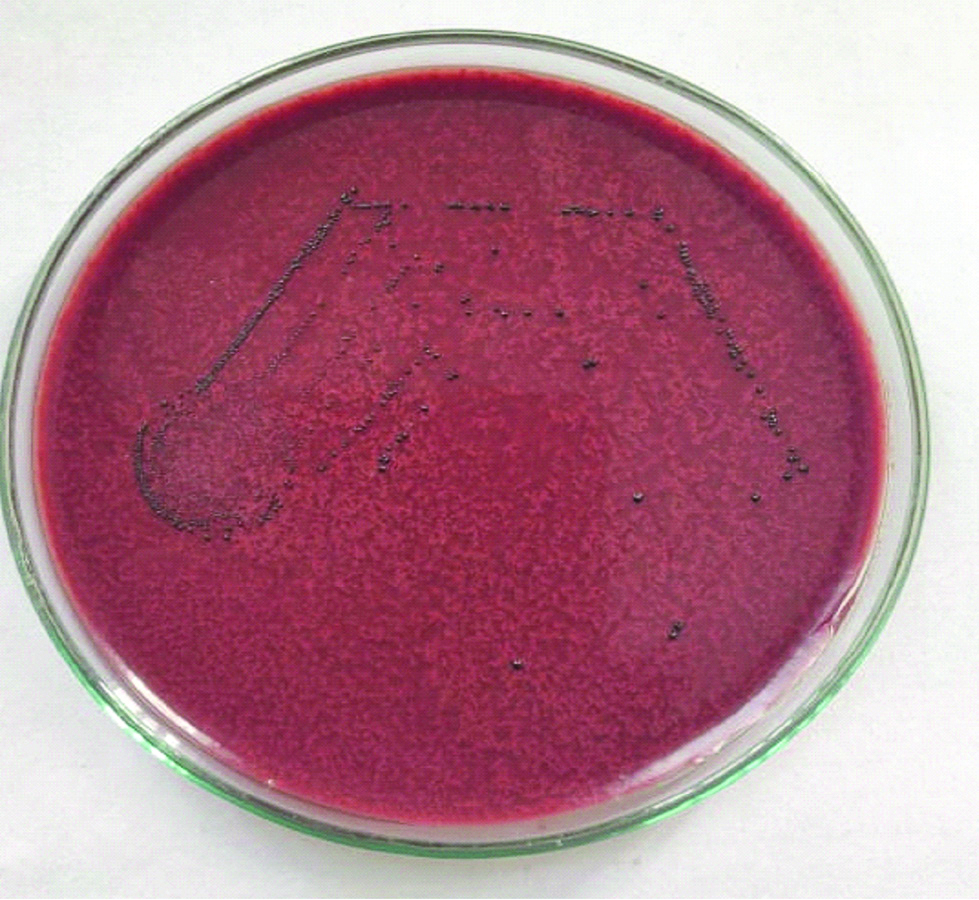
iii Tellurite reduction in Tellurite agar [Table/Fig-3]. This test is used to differentiate E.faecalis and E.faecium. E.faecalis produces black colour colonies.
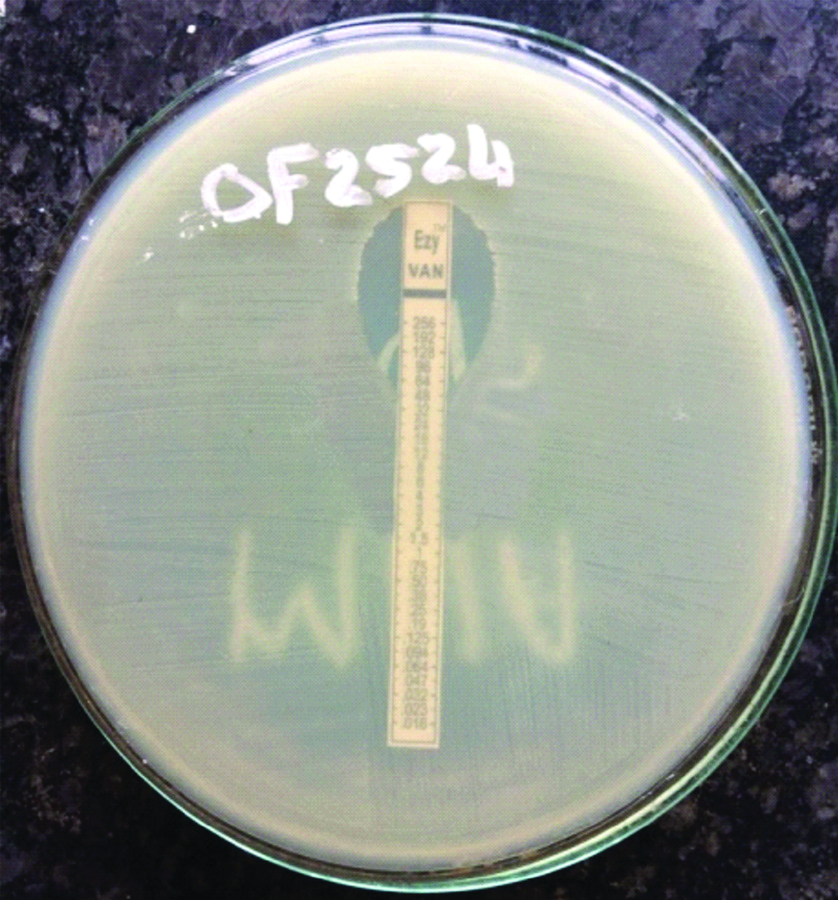
Determination of antibiotic susceptibility pattern to Pencillin (10 U), Ampicillin 10 μg), Vancomycin (30 μg), Teicoplanin (30 μg), Linezolid (30 μg), Ciprofloxacin (5 μg), Gentamicin (10 μg), High level Gentamicin (120 μg), Tetracycline (30 μg), Erythromycin (15 μg) for Enterococci were done by Kirby Bauer disc diffusion method on 5% Mueller Hinton agar as per 2017 CLSI guidelines [17]. Antibiotic susceptibility with Nitrofurantoin (300 μg) disc was done only for the urinary isolates. The minimum inhibitory concentration for Vancomycin was done by E-test method using HIMEDIA E-strips [Table/Fig-4] [17]. ATCC E. feacalis 29212 was used as the control.
Growth on potassium tellurite agar to differentiate between E.faecalis and E.faecium.
Black coloured colonies produced by Enterococcus faecalis
E-strip of vancomycin of E.faecium showing MIC=32 μg/mL.
Statistical Analysis
After coding, data was entered and analysed in Microsoft Excel version 10 and percentages of Enterococcal species and their antibiotic susceptibility was calculated.
Results
Of the 75 Enterococcal isolates, 44 (58.7%) were obtained from male patients and 31 (41.3%) were obtained from female patients. Maximum number of patients 34 (45.4%) were in the age group of 40-60 years, and least number of patients 8 (10.7%) were in age group of 20-40 years. Majority of the isolates were from pus samples 42 (56%), like diabetic foot ulcers, cellulitis, surgical site infection and abscesses [Table/Fig-5]. Out of the 18 (24%) urine samples, 8 (44.4%) were from catheterised patients. Fourteen (18.7%) Enterococci were isolated from blood and 1 (1.3%) from synovial fluid. Other samples like pleural fluid, peritoneal fluid and CSF did not yield any Enterococci. The main species isolated was E.faecalis 66.7% (50), followed by E.faecium 21.3% (16), E.raffinosus 8% (6), E.durans 2.7% (2) and E.avium 1.3% (1).
Species distribution of Enterococci in various clinical specimens.
| Samples | Total no of Enterococci n* (%) | E.faecalis n (%) | E.faecium n (%) | E.raffinosus n (%) | E.durans n (%) | E.avium n (%) |
|---|
| Pus | 42 (56%) | 25 | 8 | 6 | 2 | 1 |
| Urine | 18 (24%) | 10 | 8 | 0 | 0 | 0 |
| Blood | 14 (18.7%) | 14 | 0 | 0 | 0 | 0 |
| Synovial fluid | 1 (1.3%) | 1 | 0 | 0 | 0 | 0 |
| Total | 75 | 50 (66.7%) | 16 (21.3%) | 6 (8%) | 2 (2.7%) | 1 (1.3%) |
*n: Number
Among the 75 isolates, all strains were sensitive to Teicoplanin and Linezolid. Rate of resistance to Erythromycin was 72% (54), Penicillin 69.3% (52), Ampicillin 40% (30), Tetracycline 34.7% (26), High level Gentamicin 33.3% (25), Ciprofloxacin 30.7% (23) and Vancomycin 4% (3). The 18 urinary isolates of Enterococci showed 16.7% (3) resistance to Nitrofurantoin [Table/Fig-6]. Out of the 75 isolates, 25 (33.3%) showed High Level Gentamicin resistance, of which 15 (30%) were E.faecalis and 10 (62.5%) were E.faecium [Table/Fig-7]. None of the other strains of Enterococci showed HLGR or were VRE.
Antibiotic susceptibility testing pattern of the isolated strains.
| Antibiotics | Sensitive strains | Resistant strains |
|---|
| Number (n) | Percentage (%) | Number (n) | Percentage (%) |
|---|
| Penicillin | 23 | 30.6 | 52 | 69.3 |
| Ampicillin | 45 | 60 | 30 | 40 |
| Vancomycin | 72 | 96 | 3 | 4 |
| Gentamicin (10 μg) | 57 | 76 | 18 | 24 |
| High level- Gentamicin (120 μg) | 50 | 66.6 | 25 | 33.3 |
| Tetracycline | 49 | 65.3 | 26 | 34.7 |
| Nitrofurantoin (for 18 urinary isolates) | 15 | 83.3 | 3 | 16.7 |
| Ciprofloxacin | 52 | 69.3 | 23 | 30.7 |
| Erythromycin | 21 | 28 | 54 | 72 |
| Linezolid | 75 | 100 | 0 | 0 |
| Teicoplanin | 75 | 100 | 0 | 0 |
Vancomycin MIC was tested using E-strip for isolates which showed intermediate resistance or resistance in Disc diffusion method. By Disc diffusion method, five isolates showed intermediate resistance and two showed absolute resistance. Out of the five isolates which showed intermediate resistance by disc diffusion test, only one showed resistance by E-test. So, among the 75 isolates, 3 (4%) showed resistance to Vancomycin in E-test, 2 (4%) were E.faecalis and 1 (6.25%) was E.faecium [Table/Fig-7]. The two isolates of E.faecalis showed MIC of 32 μg/mL and 48 μg/mL respectively and one isolate of E.faecium showed MIC of 32 μg/mL. Rate of resistance was more with E.faecium compared to E.faecalis. Rate of resistance to Penicillin 14 (87.5%), Ampicillin 12 (75%), Vancomycin 1 (6.2%), High Level Gentamicin 10 (62.5%) and Ciprofloxacin 9 (56.2%) were more in E.faecium compared to E.faecalis. But only resistance for Ampicillin showed statistical significance between the two species (p-value <0.05) [Table/Fig-7].
Comparison of antibiotic resistance between E.faecalis and E.faecium.
| Antibiotics | E.faecalis (n=50) | E.faecium (n=16) | |
|---|
| No. of resistant isolates | Percentage of resistance | No. of resistant isolates | Percentage of resistance | p-value |
|---|
| Penicillin | 38 | 76 | 14 | 87.5 | 0.65 |
| Ampicillin | 18 | 36 | 12 | 75 | 0.04 |
| Vancomycin | 2 | 4 | 1 | 6.2 | 0.71 |
| Gentamicin (10 μg) | 11 | 22 | 7 | 43.8 | 0.14 |
| High level Gentamicin (120 μg) | 15 | 30 | 10 | 62.5 | 0.06 |
| Tetracycline | 18 | 36 | 8 | 50 | 0.43 |
| Nitrofurantoin (for 18 urinary isolates) | 1/10 | 10 | 2/8 | 25 | 0.43 |
| Ciprofloxacin | 14 | 28 | 9 | 56.2 | 0.09 |
| Erythromycin | 39 | 78 | 15 | 93.8 | 0.54 |
| Linezolid | 0 | 0 | 0 | 0 | |
| Teicoplanin | 0 | 0 | 0 | 0 | |
Discussion
Enterococci, a part of the normal intestinal flora produce bacteriocins, and are widely used in the food industry as probiotics or as starter cultures over the last decade [18]. Over the past 3 decades, Enterococci have emerged from relatively innocuous organism to medically important multidrug-resistant nosocomial pathogens that are considered a serious public health threat. It is due to their inherent resistance to antibiotics, ability to adhere to indwelling medical devices, and ability to survive in adverse environmental conditions.
According to the 2011-2014 National Health Care Safety network (NHSN) survey in US medical centres, 2.5% of Central Line associated Blood Stream Infections (CLABSIs) and 19.1% of surgical site infection due to Enterococci were Vancomycin resistant [19]. VRE infections are associated with additional morbidity and mortality, especially for patients with risk factors. In 2011-2012, the European CDC reported VRE prevalence of 3.6% to 31% [20]. In India, the first report of VRE was published from New Delhi, in 1999 with the present day rates ranging from 0.89% to 10% [21]. According to the 20-year SENTRY Antibiotic Surveillance programme, the frequency of VRE (VanA and VanB) only has increased in all 4 of the monitored global regions. In the early years of the SENTRY Antibiotic Surveillance programme, Program (1997-2000), VRE was uncommon (0.0%-3.0%) except in North America (10.3%); but the frequency of VRE increased in all regions through 2012 [19].
The combination of a cell-wall active agent to an Aminoglycoside is the standard of care for deep-seated Enterococcal infections due to the synergistic activity. The Clinical Laboratory Standards Institute has recommended screening of Enterococci for high-level resistance to Streptomycin and Gentamicin due to the worldwide increase of HLGR during the past. After the first report of HLGR in France in 1979, there have been reports of varied HLGR prevalence in different geographic regions 46.15% in Italy 45.5% in Brazil, 37.64% in Chicago 63% in South Korea and 62% in Delhi [22]. Due to the dynamic nature of VRE and HLGR, resistance may vary among different regions, supporting the need for ongoing surveillance and applying strict infection prevention practice.
In the present study, Enterococcal isolates were predominant in males (58.7%) as in most of the studies; which can be attributed to the increased outdoor activities, health habits, co-morbidities and risk of trauma in males [2,3,22].
In this study, Enterococcal isolates were highest from pus samples (56%) followed by urine, blood and synovial fluid. This is in contrast with many previous studies, where the highest isolates were from urine [23-25]. One of the reasons for this difference may be due to the fact that in hospitalised patients, Enterococci infections are more commonly seen in orthopaedic and surgical patients. The close proximity of anus to urethra may be one of the most probable cause for the high isolation rate of Enterococci from UTI, as Enterococci reside as commensals in GIT. Urinary catheterisation may also have contributed to higher isolation of Enterococci from urine specimens [26].
In today’s era, correct speciation is very important as the different Enterococcal species shows varied resistance to antibiotics. Out of the 75 isolates of Enterococci, E.faecalis 50 (66.7%), was the predominant species followed by E.faecium 16 (21.3%), E.raffinosus 6 (8%), E.avium 1 (1.3%), E.durans 2 (2.7%). Studies by Salem-Bekhit MM et al., in Saudi Arabia, Shanmukhappa et al., at Mysore and Haritsa KB et al., at Bangalore also found that E.faecalis was the predominant species [2,22,23]. The most putative virulence determinants associated with E. faecalis (and less frequently with E. faecium) are involved in adherence to extracellular structures and biofilm formation, which appear to be important factors for colonisation and infection.
Enterococci are intrinsically resistant to several antibiotics and also acquire resistance by conjugation using pheromone-responsive plasmids. β-lactam antibiotics such as Penicillins are not bactericidal against many strains of Enterococci; and the organism develop tolerance to Penicillins (that is, lack of killing despite growth inhibition) [9]. The treatment of choice for Enterococcal infections is considered to be Beta-lactams along with Aminoglycosides due to the synergistic effect. Recent nosocomial E. faecium isolates are found to be resistant to Ampicillin and Vancomycin, and also have high-level Aminoglycoside resistance. Currently, this has become a major concern for the clinicians worldwide, as other therapeutic options are not reliable, have toxic side effects or have not been tested in prospective randomised clinical trials. Among the 75 isolates, rate of resistance to Penicillin was 69.3% (52) and Ampicillin 40% (30) [Table/Fig-6]. All strains were sensitive to Teicoplanin and Linezolid. Similar results were seen in studies conducted by Jaiswal S et al., in Uttar Pradesh and Shah L et al., at Surat [24,27]. Linezolid can be considered as the drug of choice to treat infections with VRE; however as resistance to it has been reported, its judicious use is recommended [24,27].
In the present study, E.faecium was more resistant than E.faecalis. This is due to the fact that virulence factors like Aggregation substance (Agg), Extracellular proteins, Toxins and ability to form Biofilm is more with E.faecium [27]. High Level Gentamicin resistance detected by disc diffusion method showed resistance to 25 (33.3%) isolates out of 75. E. faecalis showed 30% resistance and E.faecium showed 62.5% resistance to High Level Gentamicin. A study conducted by Karmarkar MG et al., showed 100% HLGR in E.faecalis and 85.71% in E.faecium [28]. Study by Haritsa KB et al., showed 22.4% HLGR in E.faecalis and 30.8% in E.faecium [Table/Fig-8] [2,23,25,28]. As the mechanisms of Enterococci resistance are different for different antibiotics, Gentamicin resistance is found to be a good predictor of resistance to Aminoglycosides except Streptomycin. Hence, HLGR strains have become more challenging in infection control [29].
Rate of high level Gentamicin resistance in Enterococci from various studies [2,23,25,28].
| Authors | Year of study | Rate of resistance in E.faecalis | Rate of resistance in E.faecium |
|---|
| Karmarkar MG et al., [28] | 2004 | 100% | 85.71% |
| Salem-Bekhit MM et al., [2] | 2012 | 22.3% | 18.5% |
| Haritsa KB et al., [23] | 2014 | 22.4% | 30.8% |
| Palanisamy S et al., [25] | 2013 | 29% | 29% |
| Present study | 2017 | 30% | 62.5% |
In the present study, 3 isolates were VRE (4%) confirmed by MIC determination by E-test, out of which 2 (4%) were E.faecalis and 1 (6.25%) was E.faecium; which were isolated from urine, liver abscess and surgical site infection respectively. Various studies from different parts of India showed resistance ranging from 0.89% to 10% with a study by Karmarkar MG et al., showing as high as 28.57% [Table/Fig-9] [23,25,28,30]. VRE accounts for about 30% of Enterococcal infections, with most VRE isolates being E. faecium (>90%) [31]. Immunosuppression, serious co-morbid condition, increased hospital stay, invasive procedures, and administration of broad-spectrum antibiotics are all risk factors associated with increased VRE colonisation [32,33]. Among the 3 VRE isolates in the present study, urinary isolate of E faecalis was from a catheterised patient, E faecalis from liver abscess was isolated from an alcoholic and diabetic patient and E faecium was from the surgical wound of a patient who had undergone invasive abdominal surgery. The most common source of transmission of VRE is through the hands of health care workers, and the Society for Health Epidemiology of America has published specific guidelines to curtail this transmission [34]. Patients with bacteremia with VRE were about 2.5 times more likely to die than those with VSE (Vancomycin Sensitive Enterococci) bacteremia, indicating that the development of Vancomycin resistance is a poor prognostic sign in critically ill patients [35].
Rate of vancomycin resistance in Enterococci from various studies [23,25,28,30].
| Authors | Year of study | Rate of resistance in E.faecalis | Rate of resistance in E.faecium |
|---|
| Karmarkar MG et al., [28] | 2004 | 10% | 28.57% |
| Rahangdale VA et al., [30] | 2008 | 11.385 | 11.38% |
| Palanisamy S et al., [25] | 2013 | 0.89% | 0.89% |
| Haritsa KB et al., [23] | 2014 | 3.9% | 0% |
| Present study | 2017 | 4% | 6.25% |
Limitation(s)
There are two major limitations in this study that could be addressed in future research: first is that molecular characterisation of resistant organisms were not performed and, second is the small sample size.
Conclusion(s)
The emergence of gram-positive organisms in 1990s, as the important cause of both hospital- and community-acquired infections has warranted a revaluation of public research priorities. Multiple drug resistant strains of E. faecalis and E. faecium are progressively related to health care associated infections.
This study emphasises the need to screen for HLGR and Vancomycin in clinical isolates, active surveillance and the prompt reporting of resistance by the laboratories to prevent injudicious use of antibiotics.
The efforts of the various departments of the hospital by educating the staff, rationale use of antibiotics, early detection and reporting by laboratories and implementation of appropriate infection control measures can help in prevention and control of the spread of multi drug resistant Enterococcal infections.
*n: Number