Introduction
Several acute and chronic infections in humans can be caused by Pseudomonas aeruginosa (P.aeruginosa). Biofilm formation by these bacteria threatens health setting via increasing resistance against antimicrobial agents.
Aim
To investigate the association between lasB and nanI genes with biofilm formation in P. aeruginosa clinical isolates.
Materials and Methods
A total of 161 P. aeruginosa clinical isolates were collected. The Microtiter Plate (MTP) method was applied for biofilm formation assay. Gene amplification was conducted by PCR method with specific primers.
Results
Present study findings showed that the prevalence of lasB and the nanI were 80.7% (130/161) and 24.8% (40/161), respectively. The biofilm formation results demonstrated that 15.5% (25/161) of isolates were not able to produce biofilm. It was considerable that the prevalence of lasB in P. aeruginosa producing biofilm was higher.
Conclusion
lasB can be considered as an effective factor for biofilm formation in P. aeruginosa isolates.
Introduction
Bacteria commonly are able to attach themselves and make biofilm structures. Biofilms formation could be observed on natural, medical or industrial settings and causes adverse effects on human’s life or health [1]. Typically, in a mature biofilm, bacteria are more resistant to antimicrobial agents by different mechanisms [2].
P. aeruginosa is an important environmental organism which is capable enough to infect in human host [3]. It also causes acute and persistent infections in immunocompromised patients, especially Cystic Fibrosis (CF) individuals [4]. P. aeruginosa possesses many Virulence Factors (VFs) [5]. LasB or elastase B, encoded by lasB gene, is a bacterial metalloprotease which acts as a polysaccharide (alginate) secretion regulator, degrades elastin and surfactant protein D (SP-D). SP-D has been recognised to bacterial aggregation, alveolar macrophage function alteration and bacterial clearance regulation [6-9]. It has been shown that lasB is related to biofilm formation [6,10]. P. aeruginosa is a neuraminidase producer, which is encoded by nan gene. Some bacterial neuraminidase is responsible for airway colonisation, pathogenesis (i.e. respiratory tract infection), and biofilm formation [11,12]. Because of diverse issues by biofilm formation in bacteria, detection regulated forming genes in freely swimming cells may be a good strategy to encounter its’ formation. Therefore, we investigated the prevalence of two previously introduced biofilm regulation genes (lasB and nanI genes) and their association with biofilm formation in clinical isolates of P. aeruginosa.
Materials and Methods
Bacterial isolates and culture conditions: The present study was a cross-sectional investigation which was conducted on 161 P. aeruginosa isolates collected from clinical samples during March 2014 to February 2015 in Ilam hospitals, Iran. P.aeruginosa isolates were identified by standard biochemical tests.
Biofilm assays: The biofilm formation was evaluated by recommended protocol by Hemati et al., [5].
Gene amplification: The PCR was performed by specific primers listed in [Table/Fig-1] [13,14].
Primers sequence and PCR condition [13,14].
| Primer | Sequence | Product length | Annealing temperature | References |
|---|
| lasB forward | 5’-TTCTACCCGAAGGACTGATAC-3’ | | | |
| lasB reverse | 5’-AACACCCATGATCGCAAC-3’ | 153 bp | 55 | [13] |
| nanI forward | 5’-CGCACTATACACAGGAACACG-3’ | | | |
| nanI reverse | 5’-GCCTAGCGGAAGGATCGTCGC-3’ | 620 bp | 64 | [14] |
Statistical Analysis
The SPSS software version 16.0 with χ2 and Fisher’s-Exact programs were applied for statistical analysis. The p-values <0.05 was considered as statistical significance.
Results
PCR results: Present study demonstrated that 80.7% of isolates were positive for lasB (n=130) and 24.8% of isolates showed nanI (n=40) in their genome. Notably, it was not observed a significant association between the presences of nanI with lasB (p>0.05) [Table/Fig-2].
Electrophoresis of PCR products for nanI (lanes 1-3) and lasB (lanes 5-7); ladder 100 bp (lane 4).
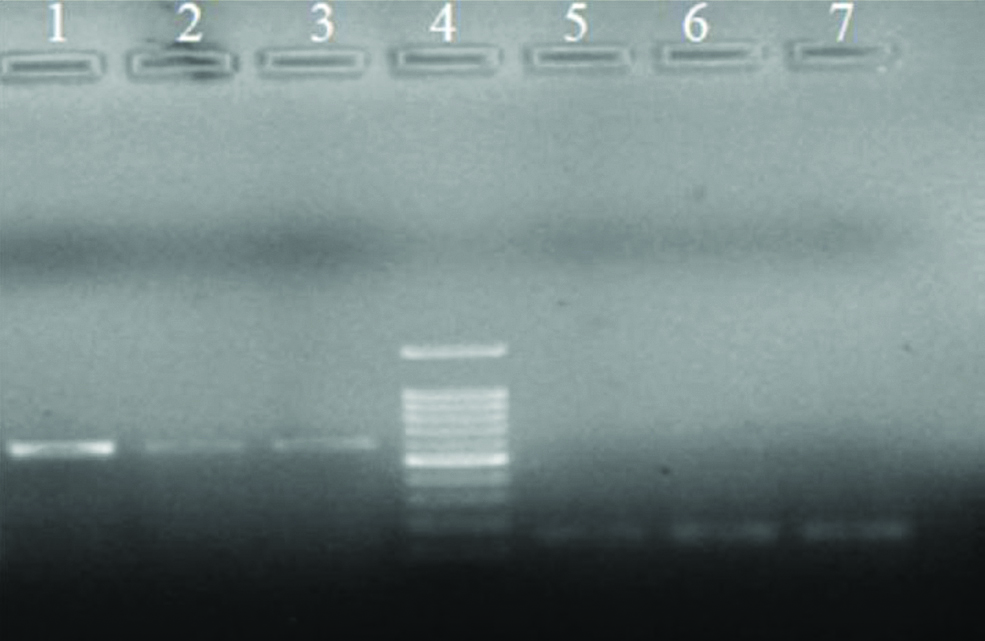
Biofilm formation results: The results showed that 84.5% (n=136) isolates were biofilm producer, while 15.5% (n=25) isolates had no ability to produce biofilm. Notably, the majority of P. aeruginosa isolates produced moderate (47.2%) and weak (24.8%) biofilm formation [Table/Fig-3]. Present study findings showed that there was a significant association between the presence of lasB and ability for biofilm formation (p=0.01), while it was not considered for nanI (p=0.102).
Biofilm formation status in different samples.
| Samples | Biofilm formation producer | Total |
|---|
| No producer | Weak producer | Moderate producer | Strong producer |
|---|
| Urine | 5 (3.1%) | 21 (13.0%) | *34 (21.1%) | 5 (3.1%) | 65 (40.4%) |
| Tracheal | 4 (2.5%) | 8 (5.0%) | *11 (6.8%) | 7 (4.3%) | 30 (18.6%) |
| Wound | 1 (0.6%) | 1 (0.6%) | *9 (5.6%) | 0 (0.0%) | 11 (6.8%) |
| Purulent exudates | 1 (0.6%) | 1 (0.6%) | *2 (1.2%) | 0 (0.0%) | 4 (2.5%) |
| Burn | 13 (8.1%) | 8 (5.0%) | *20 (12.4%) | 7 (4.3%) | 48 (29.8%) |
| CSF | *1 (0.6%) | *1 (0.6%) | 0 (0.0%) | 0 (0.0%) | 2 (1.2%) |
| Sputum | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | *1 (0.6%) | 1 (0.6%) |
| Total | 25 (15.5%) | 40 (24.8%) | 76 (47.2%) | 20 (12.4%) | 161 (100.0%) |
*Majority form of biofilm formation producer
Discussion
The pathogenicity of P. aeruginosa is multifactorial, including destroying host tissues, fighting host immune systems, biofilm formation [15,16]. P. aeruginosa is responsible for diverse human infections [17]. Here, most of the isolates were collected from urine samples (65, 40.4%). In previous published researches, maximum infection was reported from wound (45%), urinary tract (26.74%) and pulmonary tract (44%) infections [18-20].
These isolates also produces several proteolytic enzymes e.g., elastase B or pseudolysin (a Zn-metalloendopeptidase) endopeptidases enzymes that is encoded by lasB gene [21]. In the current study, lasB was presented in 80.7% of isolates. Although, Nikbin V et al., and Wolska K and Szweda P demonstrated that lasB presented in 100% and 96.8% of isolates, respectively [20,22]. Present study findings had also high frequency with lower quantity compared with mentioned results. The geographical difference and patient condition likely related to these variations.
Another important VFs in P. aeruginosa is neuraminidase, which encoded by nanI gene [20]. Its frequency (24.8%) was lower than lasB (80.7%). Mitov I et al., observed that 21.3% of isolates were positive for nanI, while lasB had 100% frequency [23]. The results of other studies were as follow: Nikbin et al.,: nanI (27.61%) and lasB (100%); Lanotte P et al.,: nanI (53%) and lasB (100%) [20,24]. Because nanI has the most prevalence in CF isolates [24], it can be probably the reason for low frequency in present study. Its frequency (89%) was also higher than elastase (84%) in CF isolates [25].
Present study finding demonstrated that 84.5% of isolates formed biofilm (12.4% strong, 47.2% moderate and 24.8% weak). Biofilm structures cause serious health problems by increasing bacterial resistance against host immune systems, biocides, antibiotics as well as various physicochemical agents [26]. Especially, biofilm formation in P. aeruginosa has been shown to play an important role in chronic infections in CF patients [27]. Wakimoto N et al., showed that 5.95%, 3.45% and 90.59% of isolates formed biofilm as strong, moderate and none or weak forms, respectively [28]. Hemati S et al., demonstrated that 19.28%, 21.4%, and 56.42% of isolates formed biofilm as weak, moderate and strong forms, while 12.85% were not able to produce biofilm formation [5]. Karimi S et al., reported that 29.3%, 38.6%, 23.3% of isolates formed strongly, moderately, and weakly biofilm respectively, while 8.6% of isolates were not able to biofilm forming [29]. Stepanovic S et al., showed that most Salmonella species and Listeria monocytogenes strains formed moderate and weak biofilm structures, respectively [30], while Borucki MK et al., reported that most L. monocytogenes strains formed strong biofilm formation [31]. However, the biofilm formation assay method, kind of medium, geographically difference of isolates can significantly be influenced on the biofilm forming in different studies. In addition, important limitation for in-vitro biofilm formation assay is that these methods are not able to exactly reflection in vivo situations [32].
It has been shown that LasB initiates biofilm formation pathway through secreted polysaccharides regulation and nucleoside diphosphate kinase (NDK) activation [33,34]. We also observed high tendency for biofilm formation among lasB positive isolates. Although it has been demonstrated that neuraminidase has relation in biofilm formation [35], no significant association between nanl (p=0.102) and biofilm formation was seen. Therefore, biofilm formation may occur by different other mechanisms [36].
Limitation(s)
Because of financial limitations, present study was conducted in a small number of molecular gene analysis that was limited to the lasB and nanI prevalence. The second is that to have better understanding of the relationship between biofilm formation and presence of lasB and nanI genes, it was better that biofilm formation compared in both bacterial wild-type and mutant-type.
Conclusion(s)
P. aeruginosa isolates have high ability for biofilim formation. Biofilim formation in these isolates depended on different mechanisms, and lasB considered as an important effective factor for biofilm formation.
*Majority form of biofilm formation producer
[1]. López D, Vlamakis H, Kolter R, BiofilmsCsh Perspect Biol 2010 2(7):a00039810.1101/cshperspect.a00039820519345 [Google Scholar] [CrossRef] [PubMed]
[2]. Jahid IK, Ha SD, A review of microbial biofilms of produce: Future challenge to food safetyFSB 2012 21(2):299-316.10.1007/s10068-012-0041-1 [Google Scholar] [CrossRef]
[3]. McKnight SL, Iglewski BH, Pesci EC, The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosaJB 2000 182(10):2702-08.10.1128/JB.182.10.2702-2708.200010781536 [Google Scholar] [CrossRef] [PubMed]
[4]. Schuster M, Greenberg EP, A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosaInt J Med 2006 296(2):73-81.10.1016/j.ijmm.2006.01.03616476569 [Google Scholar] [CrossRef] [PubMed]
[5]. Hemati S, Jalilian FA, Pakzad I, Taherikalani M, Maleki A, Karimi S, The correlation between the presence of quorum sensing, toxin-antitoxin system genes and MIC values with ability of biofilm formation in clinical isolates of Pseudomonas aeruginosaIran J Med Microbiol 2014 6(3):133-39. [Google Scholar]
[6]. Carson L, Cathcart GR, Ceri H, Walker B, Gilmore BF, Comparison of the binding specificity of two bacterial metalloproteases, LasB of Pseudomonas aeruginosa and ZapA of Proteus mirabilis, using N-alpha mercaptoamide template-based inhibitor analoguesBiochem Biophys Res Commun 2012 422(2):316-20.10.1016/j.bbrc.2012.04.15722575503 [Google Scholar] [CrossRef] [PubMed]
[7]. Brumlik M, Storey D, Post-transcriptional control of Pseudomonas aeruginosalasB expression involves the 5’ untranslated region of the mRNAFems Microbiol Lett 1998 159(2):233-39.10.1016/S0378-1097(97)00576-4 [Google Scholar] [CrossRef]
[8]. Amaya S, Pereira JA, Borkosky SA, Valdez JC, Bardón A, Arena ME, Inhibition of quorum sensing in Pseudomonas aeruginosa by sesquiterpene lactonesPhytomedicine 2012 19(13):1173-77.10.1016/j.phymed.2012.07.00322925726 [Google Scholar] [CrossRef] [PubMed]
[9]. Bleves S, Viarre V, Salacha R, Michel GP, Filloux A, Voulhoux R, Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weaponsInt J Med Microbiol 2010 300(8):534-43.10.1016/j.ijmm.2010.08.00520947426 [Google Scholar] [CrossRef] [PubMed]
[10]. Kamath S, Chen M, Chakrabarty A, Secretion of nucleoside diphosphate kinase by mucoid Pseudomonas aeruginosa 8821: Involvement of a carboxy-terminal motif in secretionJB 2000 182(13):3826-31.10.1128/JB.182.13.3826-3831.200010851000 [Google Scholar] [CrossRef] [PubMed]
[11]. Hsiao Y-S, Parker D, Ratner AJ, Prince A, Tong L, Crystal structures of respiratory pathogen neuraminidasesBiochem Biophys Res Commun 2009 380(3):4677110.1016/j.bbrc.2009.01.10819284989 [Google Scholar] [CrossRef] [PubMed]
[12]. Parker D, Soong G, Planet P, Brower J, Ratner AJ, Prince A, The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formationInfect Immun 2009 77(9):3722-30.10.1128/IAI.00228-0919564377 [Google Scholar] [CrossRef] [PubMed]
[13]. Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Bacterial neuraminidase facilitates mucosal infection by participating in biofilm productionJ Clin Invest 2006 116(8):229710.1172/JCI2792016862214 [Google Scholar] [CrossRef] [PubMed]
[14]. Petit SM-C, Lavenir R, Colinon-Dupuich C, Boukerb AM, Cholley P, Bertrand X, Lagooning of wastewaters favors dissemination of clinically relevant Pseudomonas aeruginosaRes Microbiol 2013 164(8):856-66.10.1016/j.resmic.2013.06.00723792168 [Google Scholar] [CrossRef] [PubMed]
[15]. Singh BN, Singh HB, Singh A, Singh BR, Mishra A, Nautiyal C, Lagerstroemia speciosa fruit extract modulates quorum sensing-controlled virulence factor production and biofilm formation in Pseudomonas aeruginosaMicrobiology 2012 158(2):529-38.10.1099/mic.0.052985-022117007 [Google Scholar] [CrossRef] [PubMed]
[16]. Ołdak E, Trafny EA, Secretion of proteases by Pseudomonas aeruginosa biofilms exposed to ciprofloxacinAntimicrob Agents Chemother 2005 49(8):3281-88.10.1128/AAC.49.8.3281-3288.200516048937 [Google Scholar] [CrossRef] [PubMed]
[17]. Caselli D, Cesaro S, Ziino O, Zanazzo G, Manicone R, Livadiotti S, Multidrug resistant Pseudomonas aeruginosa infection in children undergoing chemotherapy and hematopoietic stem cell transplantationHaematologica 2010 95(9):1612-15.10.3324/haematol.2009.02086720305140 [Google Scholar] [CrossRef] [PubMed]
[18]. Troillet N, Samore MH, Carmeli Y, Imipenem-resistant Pseudomonas aeruginosa: Risk factors and antibiotic susceptibility patternsClin Infect Dis 1997 25(5):1094-98.10.1086/5160929402364 [Google Scholar] [CrossRef] [PubMed]
[19]. Tavajjohi Z, Moniri R, Khorshidi A, Detection and characterization of multidrug resistance and extended-spectrum-beta-lactamase-producing (ESBLS) Pseudomonas aeruginosa isolates in teaching hospitalAfrican J Microbiol Res 2011 5(20):3223-28.10.5897/AJMR11.260 [Google Scholar] [CrossRef]
[20]. Nikbin V, Aslani MM, Sharafi Z, Hashemipour M, Shahcheraghi F, Ebrahimipour G, Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious originsIran J Med Microbiol 2012 4(3):118 [Google Scholar]
[21]. Ghorbel-Bellaaj O, Hayet BK, Bayoudh A, Younes I, Hmidet N, Jellouli K, Pseudomonas aeruginosa A2 elastase: Purification, characterization and biotechnological applicationsInt J Biol Macromol 2012 50(3):679-86.10.1016/j.ijbiomac.2012.01.03822326423 [Google Scholar] [CrossRef] [PubMed]
[22]. Wolska K, Szweda P, Genetic features of clinical Pseudomonas aeruginosa strainsPol J Microbiol 2009 58(3):255-60. [Google Scholar]
[23]. Mitov I, Strateva T, Markova B, Prevalence of virulence genes among Bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosaBraz J Microbiol 2010 41(3):588-95.10.1590/S1517-8382201000030000824031533 [Google Scholar] [CrossRef] [PubMed]
[24]. Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other originsJ Med Microbiol 2004 53(1):73-81.10.1099/jmm.0.05324-014663109 [Google Scholar] [CrossRef] [PubMed]
[25]. Lanotte P, Mereghetti L, Lejeune B, Massicot P, Quentin R, Pseudomonas aeruginosa and cystic fibrosis: Correlation between exoenzyme production and Patient’s clinical statePediatr Pulmonol 2003 36(5):405-12.10.1002/ppul.1038014520723 [Google Scholar] [CrossRef] [PubMed]
[26]. Chavant P, Gaillard-Martinie B, Talon R, Hébraud M, Bernardi T, A new device for rapid evaluation of biofilm formation potential by bacteriaJ Microbiol Meth 2007 68(3):605-12.2379216817218029 [Google Scholar] [CrossRef] [PubMed]
[27]. Musk DJ, Banko DA, Hergenrother PJ, Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosaChem Biol 2005 12(7):789-96.10.1016/j.chembiol.2005.05.00716039526 [Google Scholar] [CrossRef] [PubMed]
[28]. Wakimoto N, Nishi J, Sheikh J, Nataro JP, Sarantuya J, Iwashita M, Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coliAm J Trop Med Hyg 2004 71(5):687-90.10.4269/ajtmh.2004.71.68715569806 [Google Scholar] [CrossRef] [PubMed]
[29]. Karimi S, Ghafourian S, Kalani MT, Jalilian FA, Hemati S, Sadeghifard N, Association between toxin-antitoxin systems and biofilm formationJundishapur J Microbiol 2015 8(1)10.5812/jjm.1454025789127 [Google Scholar] [CrossRef] [PubMed]
[30]. Stepanovic S, cirkovic I, Ranin L, Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surfaceLett Appl Microbiol 2004 38(5):428-32.10.1111/j.1472-765X.2004.01513.x15059216 [Google Scholar] [CrossRef] [PubMed]
[31]. Borucki MK, Peppin JD, White D, Loge F, Call DR, Variation in biofilm formation among strains of Listeria monocytogenesAppl Environ Microbiol 2003 69(12):7336-42.10.1128/AEM.69.12.7336-7342.200314660383 [Google Scholar] [CrossRef] [PubMed]
[32]. Stepanovic S, Vukovic D, Hola V, Bonaventura GD, Djukic S, Cirkovic I, Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococciAPMIS 2007 115(8):891-99.10.1111/j.1600-0463.2007.apm_630.x17696944 [Google Scholar] [CrossRef] [PubMed]
[33]. Sharma G, Rao S, Bansal A, Dang S, Gupta S, Gabrani R, Pseudomonas aeruginosa biofilm: potential therapeutic targetsBiologicals 2014 42(1):01-07.10.1016/j.biologicals.2013.11.00124309094 [Google Scholar] [CrossRef] [PubMed]
[34]. Cathcart GR, Gilmore BF, Greer B, Harriott P, Walker B, Inhibitor profiling of the Pseudomonas aeruginosa virulence factor LasB using N-alpha mercaptoamide template-based inhibitorsBioorg Med Chem Lett 2009 19(21):6230-32.10.1016/j.bmcl.2009.08.09919773163 [Google Scholar] [CrossRef] [PubMed]
[35]. Xu G, Ryan C, Kiefel MJ, Wilson JC, Taylor GL, Structural studies on the Pseudomonas aeruginosa sialidase-like enzyme PA2794 suggest substrate and mechanistic variationsJ Mol Biol 2009 386(3):828-40.10.1016/j.jmb.2008.12.08419166860 [Google Scholar] [CrossRef] [PubMed]
[36]. Ghafourian S, Raftari M, Sadeghifard N, Sekawi Z, Toxin-antitoxin systems: Classification, biological function and application in biotechnologyHorizon Scientific Press 2014 [Google Scholar]