CMV is a quiet invader for immuno-competent persons with no or few symptoms but is considered threat to immuno-compromised individuals such as AIDS patients, allograft organ transplant recipients, etc. The sero-prevalence of CMV infection ranges from 55% to 80% in developed countries to as high as 90% in developing countries [1]. For the prevention of CMV disease in immuno-suppressed patients, pre-emptive antiviral therapy and a strict follow-up of CMV infection are the standard of care over the past decades. Virological monitoring of viral replication for timely antiviral treatment is also necessary to prevent the disease progression [2].
Ganciclovir, which is considered as the inhibitor of Herpes virus family is successfully used to treat active CMV. But the duration of antiviral treatment therapy to avoid recurrence after the treatment is still a dangerous clinical question. Thus, regular monitoring of viral loads for CMV infection is advisable and treatment with antiviral therapy until viremia is no longer detectable are recommended. Though antiviral treatment reduces the mortality and morbidity rates, for identifying the presence of HCMV infection, the availability of highly sensitive and accurate laboratory tests are of great significance [3].
Conventional PCR and non-PCR methods have been largely replaced by new molecular technologies based on the quantitative real-time Polymerase Chain Reaction (PCR) with very acceptable clinical results for quantifying the viral load and decreasing the chances of contamination by reacting in a closed system [4]. More sensitive assays give opportunity for the earlier detection of CMV infection which in turn improves the ability of pre-emptive treatment strategies. The positive rate of HCMV infection is also affected by different blood compartments (e.g., Whole blood vs. Plasma) apart from detection assays. Currently, to begin the pre-emptive anti-CMV therapy, there is no universally accepted viral load threshold limit. They are clinically validated in certain settings but all refer to a specific biologic matrix [4,5]. Completely automated system from nucleic acid extraction to real-time PCR, consumes less time and allows better standardisation of the procedure.
Continuation of treatment is recommended by current consensus guidelines because constant viremia at end of treatment is linked with elevated recurrence rates and delayed disease resolution [6]. For monitoring therapeutic response in patients with CMV disease, there would be more sensitive detection of residual viremia which is clinically useful. Viral load kinetics on both PL and WB were studied and compared for the detection of active CMV infection obtained by using real time PCR assay on immuno-suppressed patients. Diagnostic and prognostic information on CMV infection is provided by both WB and PL. This hypothesis would provide higher sensitivity detection method that would allow better prediction of recurrence of CMV infection. The post-treatments were assessed to test by early viral kinetics and end-of-treatment viral clearance.
The current work hence has been undertaken to detect the number of cases affected in the referral cases with respect to age, viral load, gender and follow-up of about 22 months (March 2017 to December 2018). For detecting HCMV infection in high risk patients to set up a highly sensitive assay applicable was also the major aim of present study.
Materials and Methods
A prospective study was conducted in which Bone Marrow transplant recipients who gave informed consent, were enrolled from March 2017 to December 2018. This project was supported/approved by Gujarat University (Ahmedabad) Human Ethical Committee (GUHEC-001/2015) for investigation. A total of 1021 referral patient samples with 2112 number of follow-ups were included for the CMV real-time PCR detection. Among these, 315 were paediatric patients while the rest 706 were in adult age group [Table/Fig-1]. EDTA-blood samples were collected after transplantation as a part of routine follow-up. Both Whole blood (1 mL) and plasma (1 mL) were recovered.
Patient’s characteristics with statistics.
| Patient’s screened | 1021 |
| No follow-ups patients | 760 (74.43%) |
| Patients detected with follow-ups | 261 (25.56%) |
| Total no. of follow-ups | 2112 |
| Lowest follow-up | 1 |
| Highest follow-up | 25 |
| Paediatric patients | 315 |
| Adult patients | 706 |
| Average time duration of follow-up (days) | 74.55 (7-75 days interval) |
DNA Extraction
DNA was extracted from 200 μL of plasma and whole blood stored at -20°C respectively using automated QIAsymphony DNA mini kit (Qiagen, Germany) according to manufacturer’s instructions and diluted in 50 μL nuclease free water. The DNA was stored in -80°C till further use for all samples.
Real-Time PCR
Amplification was performed in 20 μL reaction mixture containing: 1X Roche ROX Universal PCR Master Mix, 1 uL of CMV Taqman assay (Applied Biosystems) and 2 uL of extracted DNA. As an internal control, we used the Taqman Exogenous Internal Positive Control Reagents kit with 1X IPC Mix (Primers and Taqman probe labelled with VIC) and 1X IPC DNA. All the reagents were obtained from Applied Bio-systems (Foster City, CA, USA) and cycled according to following instructions: 95°C for 10 minutes, followed by 45 two-step cycles of 95°C for 15 seconds and 60°C for 60 seconds and a final extension at 37°C for 30 seconds. The CMV viral load was measured as the IU/mL of DNA (Roche LightCycler 96 real-time PCR system).
Preparation of in House Standard on Digital PCR and Performance of Real-Time
CMV standard panel (AcroMetrix Corp., Benicia, CA) were used to produce in-house plasma standard that was validated on Quant studio 3D digital PCR. The levels of plasma CMV DNA viral load were determined using standards that were calculated with Quant studio 3D digital PCR (RT PCR) which is the most precise technique [7].
The standards were serially diluted in the negative plasma to generate a standard curve covering 9-Log10 dynamic linearity over the range of 7×109 to 7×101 IU/mL and were tested in triplicate along with the negative controls. Intra-assay and inter-assay variation were determined by testing the reproducibility of three CMV sero-positive samples with viral load of 2 Log10, 2.50 Log10 and 3 Log10 IU/mL, in eight replicates, in a single run for three days. Reproducibility was decided by calculating the Coefficient of Variation (CV%) [7].
The linear dynamic range of the CMV real-time PCR assay was assessed using an 9-log10 dilution series over the entire range of 7×101 to 7×109 IU/mL. Clinical specificity was determined using 25 CMV sero-negative and 50 sero-positive samples (sero-positives covering from 3 Log10 IU/mL to around 8 Log10 IU/mL of CMV concentration), resulting in a clinical specificity of 100%. Further, CMV real-time PCR assay showed no cross-reactivity with the samples of HIV, HCV, HBV and EBV, respectively. The mean intra-assay and inter-assay precision was less than around 5% and 10% respectively.
Statistical Analysis
Validation of qPCR was analysed by scatter and linear regression. For reproducibility analyses, viral loads were summarised by means of Standard Error (SE) and Coefficient of Variation (CV). Scatter plots were performed to assess the agreement between the real time PCR and Digital PCR. Statistical significance was set at p<0.05.
Results
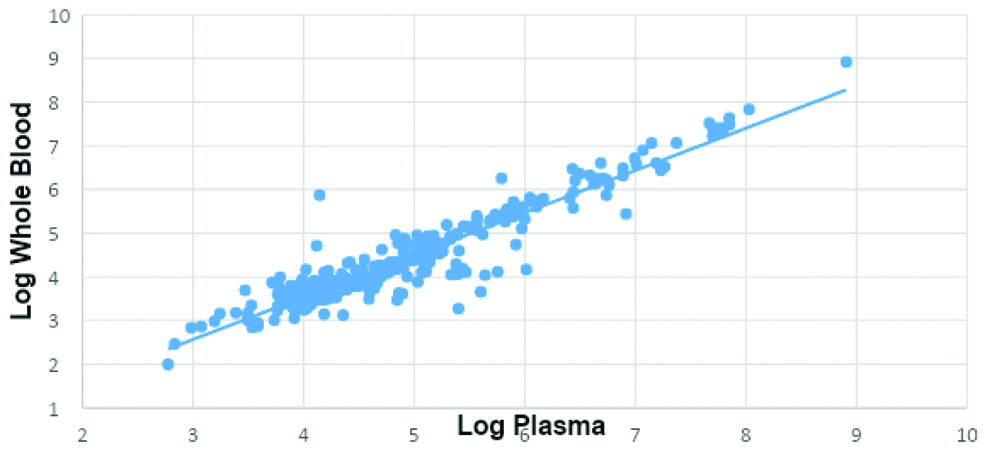
In present study, a good correlation (R2=0.89) between WB and PL for the total number of referral cases of CMV infected Bone Marrow Transplant (BMT) recipients for both the groups (Paediatric and Adult) was observed [Table/Fig-2]. Out of 2112 follow-ups of 261 patients, there were 1676 (79.35%) of follow-ups of patients which were neither detected from whole blood nor plasma sample types and were given target negative samples. Other 335 follow-ups (15.86%) were detected from both whole blood and plasma samples. There were a total 101 patients having undetected viral load from plasma but detected from whole blood which comprises about 4.78% total population [Table/Fig-3].
Whole blood versus plasma CMV viral Load (Log10 copies/mL).
Detection of CMV in WB and PLASMA among all follows-up patient.
| Whole blood detected | Whole blood not detected |
|---|
| Plasma detected | 335 (15.86%) | 0 (0%) |
| Plasma not detected | 101 (4.78%)* | 1676 (79.35%) |
*Viral load=Log10 3 IU/mL obtained by Ct values
Among 171 paediatric patients, with 1260 follow-up studies, 128 follow-ups were positive in both whole blood and plasma while 72 follow-ups were positive in WB while negative in PL. Study among 90 adults patients with 850 follow-up studies, 192 were both WB and PL positive [Table/Fig-4].
Incidence of CMV infection during follow-up in adult and paediatric groups.
| Age groups (yrs) | Paediatric (0-17) | Adult (18 onwards) |
|---|
| Total no of follow-up patient | 171 | 90 |
| Total follow-ups* | 1260 | 850 |
| Paediatric patients |
| WB detected | WB not-detected |
| Plasma detected | 128 (10.64%) | 0 (0%) |
| Plasma not-detected | 72 (5.98%) | 1060 (83.37%) |
| Adults patients |
| WB detected | WB not-detected |
| Plasma detected | 192 (22.66%) | 0 (0%) |
| Plasma Not-Detected | 30 (3.54%) | 628 (73.78%) |
*Two cases were dropped due to lack of age details in Test Requisition Form (TRF), making the number 2110
Males were found to be more affected in both (58.73%, 65.72%) groups. Thus, 64% of the total population were males affected by CMV infection and Male to female ratio was 1.7: 1.0 [Table/Fig-5].
Gender comparison in adult and paediatric groups.
| Age groups | Paediatric | Adult |
|---|
| Gender | Male | Female | Male | Female |
|---|
| Total no. of patients | 185 (58.73%) | 130 (41.26%) | 464 (65.72%) | 242 (34.27%) |
| Total cases (1021) | 315 (30.85%) | 706 (69.1%) |
There were 760 (74.43%) patients which had no follow-up studies. Thus the follow-up studies were analysed from the number of patients with lowest i.e., 1 time follow-up to highest i.e., 25 follow-ups. This shows as the number of follow-up increases, the patient size decreases along with the viral load. Few cases of patients whose viral load value were detected from whole blood (4.78%) but have become negative from their plasma samples. The average viral load pattern from both whole blood (positive/negative) and plasma (positive/negative) is shown in [Table/Fig-6]. No significant difference was detected in viral load of both components of blood.
Comparision of cases with follow-ups, viral load in Whole Blood (WB) and Plasma (PL).
| Follow-up | No. of patients | No. of follow-ups | Whole blood | Plasma | Whole blood viral load (log) | Plasma viral load (log) |
|---|
| Positive | Negative | Positive | Negative |
|---|
| No follow-up | 760 | 760 | 153 | 607 | 126 | 634 | 4.79±24.14 | 4.60±24.07 |
| 2 | 93 | 186 | 64 | 122 | 59 | 127 | 4.99±22.92 | 4.58±24.70 |
| 3 | 25 | 75 | 21 | 54 | 17 | 58 | 4.38±20.82 | 4.15±21.30 |
| 4 | 25 | 100 | 26 | 74 | 19 | 81 | 4.55±30.62 | 4.64±29.43 |
| 5 | 19 | 95 | 16 | 79 | 11 | 84 | 4.51±25.31 | 4.63±25.84 |
| 6 | 21 | 126 | 19 | 107 | 15 | 111 | 4.29±21.38 | 4.09±22.12 |
| 7 | 23 | 161 | 22 | 139 | 17 | 144 | 4.00±15.74 | 3.65±11.45 |
| 8 | 16 | 128 | 19 | 109 | 8 | 120 | 3.62±9.63 | 3.60±06.13 |
| 9 | 12 | 108 | 21 | 87 | 15 | 93 | 3.89±16.94 | 3.80±24.76 |
| 10 | 8 | 80 | 10 | 70 | 9 | 71 | 3.99±18.20 | 3.60±19.16 |
| 11 | 4 | 44 | 10 | 34 | 7 | 37 | 4.46±21.46 | 4.19±13.61 |
| 12 | 4 | 48 | 13 | 35 | 5 | 43 | 3.77±18.7 | 4.02±4.87 |
| 15 | 4 | 60 | 9 | 51 | 7 | 53 | 4.83±24.78 | 4.19±17.08 |
| 17 | 2 | 34 | 8 | 26 | 4 | 30 | 3.84±12.42 | 3.69±10.55 |
| 18 | 1 | 18 | 4 | 14 | 1 | 17 | 3.91±8.24 | 4.16 |
| 20 | 1 | 20 | 4 | 16 | 4 | 16 | 4.46±17.39 | 4.16±19.55 |
| 21 | 1 | 21 | 12 | 9 | 7 | 14 | 4.39±20.00 | 3.98±7.53 |
| 23 | 1 | 23 | 2 | 21 | 2 | 21 | 4.26±2.49 | 3.58±4.06 |
| 25 | 1 | 25 | 3 | 22 | 2 | 23 | 3.94±7.76 | 3.52±6.81 |
| Total | 1021 | 2112 | 436 | 1676 | 335 | 1777 | - | - |
In most patients (436), whole blood viral loads were lesser than 1-log which is insignificant (p=0.8963) as compared to plasma patients (335). The tendency further indicates that lower viral load was detected in PL comparatively to whole blood which was not significant [Table/Fig-6,7].
Comparison between WB and plasma in log10 Copies/mL by Real-Time PCR (Digital) for CMV monitoring.
| Viral load (log) | No. of patients in WB | Average | No. of patients in plasma | Average |
|---|
| Log 8 | 2 | 8.46 | 2 | 8.37 |
| Log 7 | 14 | 7.47 | 14 | 7.08 |
| Log 6 | 27 | 6.48 | 27 | 5.96 |
| Log 5 | 82 | 5.40 | 82 | 4.79 |
| Log 4 | 150 | 4.41 | 150 | 3.90 |
| Log 3 | 147 | 3.59 | 58 | 3.43 |
| Log 2 | 14 | 2.82 | 2 | 2.43 |
| TOTAL (Mean±SE)* | 436 | 5.52±37.40 | 335 | 5.14±41.08 |
*p=0.8963 (by Student’s t-test)
Discussion
This study was carried out in 1021 cases to monitor for CMV infection and its prognostic value in clinical condition. Whole blood reflects both cell associated and plasma free virus, thus considered as the most appropriate blood compartment. But plasma is simpler than WB, and supported by others [1,8,9]. Numerous authors have directed monitoring of CMV loads using WB, PL and Leucocytes in BMT recipients of AIDS and leukaemia cases [3] using PCR. The study also revealed more males were affected by CMV infection in both groups i.e., adult and paediatric. This could be because of the reason that males were supposed to be active and exposed to environmental factors more as compared to females. Geographical distribution of cases is also important for the cause of probable infection of CMV. This study only supports the follow-up type without treatment in the literature. These studies have analysed the infection with an interval of 7, 15, 30 days. Thus this cohort revealed that as the number of follow-ups in patients infected with CMV increases, the viral load value decreases. Such studies have also been done using antiviral therapy (drugs) as well as interval timings to reduce this infection [7]. These studies are well supportive that patients analysed with low viral load in whole blood may not have been a good predictor for antiviral treatment [1,7]. Also, plasma is considered as a significant predictor of recurrence of the disease than the whole blood by PCR technique. Thus, the present study indicated that the plasma samples are better than whole blood for CMV infection in our referral cases though the log values of CMV was not significant. Quantification of CMV has been useful in clinical contents, like virological surveillance of transplant recipients and monitor antiviral therapy [9-14]. These assays are very useful in these conditions too [10,15,16]. The Real time PCR technically measures quantitative level of HCMV DNA that is useful for predicting disease and monitoring therapy [10]. Since CMV is cell associated, its quantification in acellular fractions of the blood is also done, which is of note worthy [10,17,18]. Selection of the specimen depends on method and clinical settings [10,12,19]. The use of leukocytes may be more appropriate in situations where the viral DNA load is low or when pre-emptive therapy is envisaged where there is rapid progression of HCMV disease [10,20]. In this study, we used whole EDTA blood in order to recover viral DNA from both cellular and acellular fractions. An internal control was successfully amplified from all DNA samples extracted, indicating that nucleic acids extracted from whole EDTA blood were free of amplification inhibitors.
By using automated processes, DNA extraction can give better reproducibility, accurate CMV analysis (R=0.87, p<0.001), with improved sensitivity and increased speed of sample analyses. Such PCR assays brought about significantly more sensitivity than pp65 antigenemia and blood cultures to quantify CMV by primer with recent technologies DNA extraction techniques. Similar results were documented by others who had studied CMV infected blood compartments [7,21]. Studies by Spector SA et al., [21,22] have shown that detection of CMV DNA in blood plasma by PCR may associate with disease superior than other assays [23]. Using a whole-blood real-time PCR, measurement of CMV viral load in whole blood can increase the test sensitivity. But other significant clinical advantages such as prediction of recurrence of CMV viremia or disease compared to those with viral load detection in plasma were not obtained [7]. CMV is highly cell associated, and leukocytes have high viral load than WB and PL [23]. Plasma Viral Loads (VL) has modest clinical utility of CMV disease [3,23]. On the other hand, when CMV is detected in plasma it reflects on active viral replication with viral realise into it [24]. However, rest all believe that no difference is obtained by using WB, PL and other blood components, depending upon the technique applied, kit used and other factors [9,10,22,25-27]. We also support the same contention that WB and PL monitoring of HCMV makes not much difference, as detected in this study.
Further the results of the present study suggested RT PCR to be a better technique for detection of the viral disease for clinical use comparatively. Same suggestion was reported by other workers who utilised RT PCR system [12]. They suggested that a suitable sensitive test that would confirm CMV infection which is of clinical importance in CMV testing. Hence, an increased analytical sensitivity of the PCR assays lead to a lower clinical specificity. Few other studies concluded that CMV DNA detected by PCR in plasma was comparable to that in leukocytes or WB with PP65 antigenemia assay [21]. However, the molecular quantitative DNA assays provide earlier detection of CMV virus which in turn results in increased sensitivity [22]. So, RT PCR is able to identify the disease appropriately for follow-up studies.
Limitation(s)
This study is limited to WB and PL, other coordinates need to be analysed for better comparison. Digital PCR gives more accurate results than RT-PCR as it gives absolute quantification, so more number of samples with low viral copies should be studied on Digital PCR as well as for better justification of the results with treatment in future.
Conclusion(s)
CMV infection monitoring results in more or less similar viral loads in both the blood component matrices i.e., whole blood and plasma using real-time PCR assay. Further, PL is less contaminated due to diluents and amplification inhibitors and thereby could be more preferred matrix for the CMV infection monitoring. Quantstudio 3D digital PCR used for the preparation of in-house standards (avoiding the purchase of external standards) helped in making the real-time PCR assay cheaper and accurate for the detection of CMV in patients.
*Viral load=Log10 3 IU/mL obtained by Ct values
*Two cases were dropped due to lack of age details in Test Requisition Form (TRF), making the number 2110
*p=0.8963 (by Student’s t-test)