The OSCC is one of the most common carcinomas worldwide and usually involves multi-step carcinogenesis [1]. Many oncogenes (CCND1, EGFR, RAS, VEGF and STAT3) and tumour suppressor genes (p53, CDKN2A and Rb) have been implicated in oral cancer [2]. Tissue biopsy is gold standard and accurate diagnostic test, but it is invasive, time-consuming, having limitations temporally and spatially and provide information of a single region of a heterogeneous tumour [3]. Extensive researches are focusing on novel, non-invasive methods for the diagnosis that may provide comprehensive picture of the tumour genomic architecture for real time monitoring tumour evolution and therapeutic response [4,5]. Circulating free DNA (cfDNA) analysis commonly referred to as “liquid biopsy”, which provides information about molecular composition and presence of tumour without the need for direct tumour biopsy [6,7]. It is based on the detection of exosomes, proteins, ctDNA, circulating tumour RNA (ctRNA) and Circulating Tumour Cells (CTCs) [8]. ctDNA originates from leukocytes that undergo apoptosis and also shed from dead healthy cells, normal cells or from cancer cells [9,10]. With variable proportions, the sum of tumour and normal ctDNA gives total cfDNA. Liquid biopsies can provide extensive informations on primary and metastatic tumours at successive time points, that will help to assess the tumour burden, detecting recurrence or resistance early and helping management planning accordingly [11,12].
To assess ctDNA, both short fragment (ALU 115) and long fragment (ALU 247) of ALU repeats can be amplified and quantified. DNA integrity can be calculated as a ratio of longer to shorter DNA fragments; a higher ratio favours presence of malignancy. The presence of cancer specific genomic alterations (for example, point mutations) and difference in DNA fragment base pair length allows the differentiation between ctDNA and DNA from normal healthy cells [13]. An additional discriminating factor is the cellular apoptosis creates shorter DNA fragments (100 to 200 base pair), whereas necrosis, due to more irregular digestion, creates larger fragments (many kilo-base pair) [13,14]. Many tumour characteristics are associated with ctDNA concentrations, such as overall size, staging, vascularity, cellular turnover, location, and response to the treatment [15]. The concentration of circulating DNA in healthy patients is generally in the region of <5 ng/mL, however, cancer patients have raised levels of several hundred ng/mL [13,14]. ctDNA may also be useful for the monitoring of pre-malignant lesions in which the best course of treatment is still debated or when a biopsy may miss potential malignancy in a severely dysplastic lesion [15]. In the post-treatment phase, the high sensitivity of ctDNA poses a real opportunity for the first biomarker in HNSCC to assess for residual disease or loco-regional recurrence. Different technologies for ctDNA detection have been developed allowing it to be analysed from the level of a point mutation to that of the entire genome [16]. Classical methods of analysing ctDNA include quantitative real-time PCR, fluorescent assays, and spectrophotometric strategies, next-generation sequencing [4]. ctDNA analysis could improve the precision of risk stratification and identify patients with residual or recurrent disease. The Primary Outcome Measures of the presenting study is to assess changes in ctDNA values in relation to response to therapy. The Secondary Outcome Measures is the association between ctDNA and clinico-pathological parameters. The objective was to assess ctDNA as response assessment tool and as a clinico-pathological parameter, complementing the diagnosis of oral cancer.
Materials and Methods
This prospective study was conducted at tertiary care cancer centre, Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow, India, after taking ethical approval (IEC number- 30/16) from May 2017 to November 2018.
After proper consent, 25 patients with Eastern Cooperative Oncology Group performance status 0-2 having resectable treatment naive oral cavity cancer were included.
Patients with second malignancy, unwilling to take active treatment, co-existing medical condition likely to interfere with laboratory results were excluded.
The analysed parameters were sex, age, race, tobacco chewing and smoking history, familial history of cancer, tumour site, biopsy report, h/o of treatment, recurrence and development of second primary tumour during follow-up. ctDNA testing was done on pre-surgical blood sample collected one day before surgery, post-recovery within 1-2 months and at recurrence. All resection specimens were analysed histopathologically. Follow-up was done at 1 week and 1-2 months after surgery with clinical examination. Further follow-up was done for development of recurrence. In case of any recurrence, ctDNA levels were repeated along with the other investigations for diagnosis. Sample size is time dependent as per the prospective study.
Experimental Technique
An amount of 3.5 mL of peripheral blood was collected in silica gel vials. Serum separation and ctDNA isolation were performed. Purified ctDNA was frozen at -80°C until further processing. DNA reference standards S1 to S4 (10 ng-0.01 pg) were prepared using Standard TaqMan Control Human Genomic DNA (Applied Biosystem, USA) with concentration of 10 ng/μL. The standard 1 ng (S2), 0.1 ng (S3) and 0.01 ng (S4) were prepared by serial dilution of stock 10 ng (S1) in nuclease free water. Both ALU115 and ALU247 primer sets were given standard curves. The ALU 115 primer set represented the total amount of ctDNA (amplify both apoptotic and non-apoptotic DNA fragment). ALU 247 primer set amplify only non-apoptotic DNA fragments. The ratio of ALU247-qPCR/ALU115-qPCR denoted ctDNA integrity index. The forward and reverse sequences of the ALU115 primers were 5-CCTGAGGTCAGGAGTTCGAG-3 and 5-CCCGAGTAGCTGGGATTACA-3; and of the ALU247 primers were 5-GTGGCTCACGCCTGTAATC-3 and 5-CAGGCTG GAGTGCAGTGG-3; respectively. Each 20 μL reaction mixture for each of ALU-qPCR consisted of 2 μL of DNA, 0.5 μM each of forward primer and reverse primer (ALU115 or ALU247), 10 μL of SYBR Green Supermix (Applied Biosystems, USA), and the volume was adjusted by nuclease free water. qPCR was performed on CF X96 Real-Time PCR system (Bio-Rad Laboratories, Hercules, CA, USA) with thermal cycling conditions of first denaturation at 95°C for 9 minutes, followed by 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds. To confirm the generation, single specific PCR product melting curve analysis was performed from 65°C to 95°C (increment 0.5°C every 30 seconds) at the end of each reaction. All samples were made in duplicates and a negative control (without DNA) was set in each run. Mean values were calculated from duplicate reactions [Table/Fig-1].
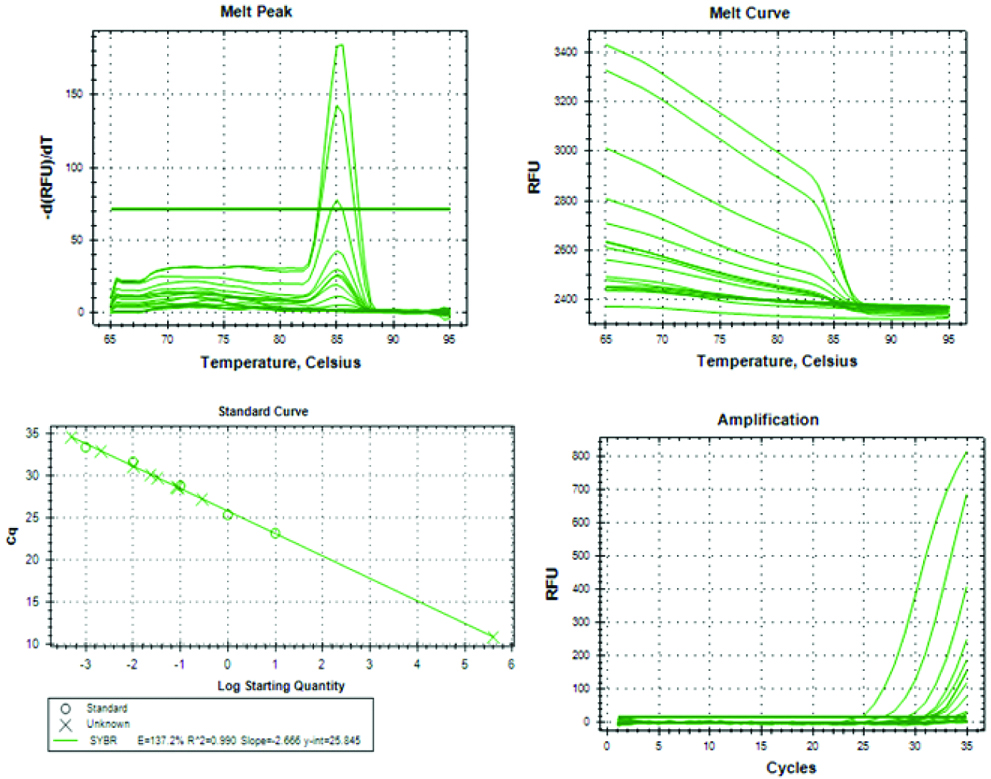
Statistical Analysis
The predictive value of ctDNA for DFS/PFS was evaluated by Cox proportional model. Association between circulating tumour DNA with clinico-pathological parameters predicting response and prognosticating disease was done in terms of univariate and multivariate analysis for cfDNA and study variables. STATA version 12.0 was used.
Results
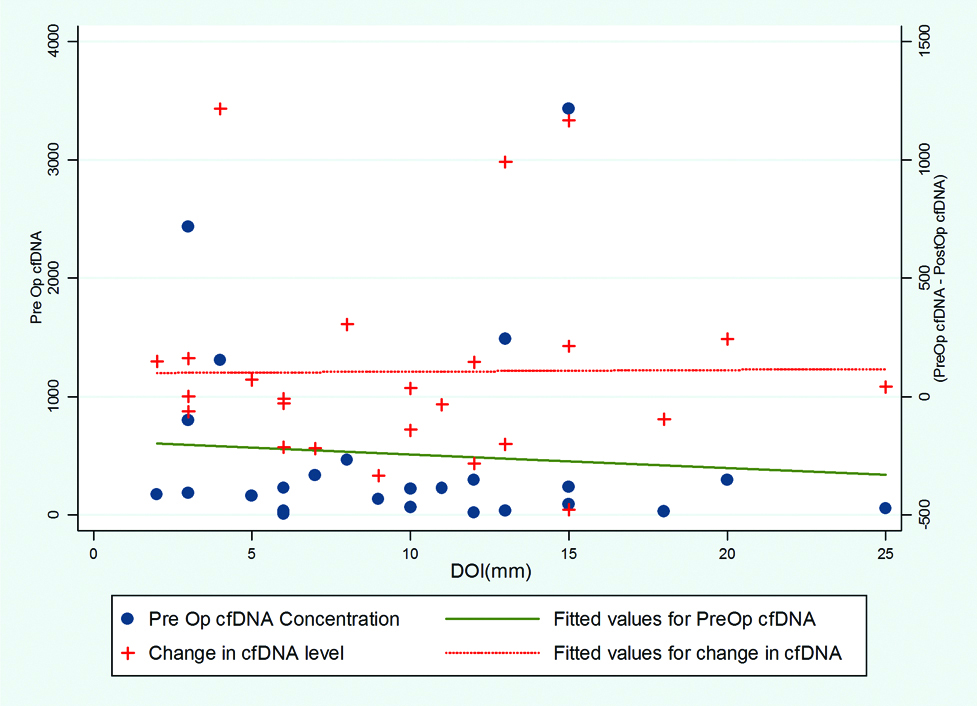
Twenty-five cases of biopsy proven OSCC were studied and recorded all demographic, personnel and disease related data. The data is compared in relation to pre-operative ctDNA levels, change of ctDNA levels and its relation to clinico-pathologic parameters. A non-significant trend of higher ctDNA among the patient with age ≤40 years and female sex was noted. Similarly, higher values were noted among patients presented with shorter duration (≤3 months) of illness (mean Pre-operative ctDNA: 628.99 and 362.19, respectively). ctDNA levels were lower among chronic smokers and tobacco chewers. The correlation between subsites of oral cavity and ctDNA levels cannot be analysed due to small sample size. The present authors noted higher ctDNA levels among lower tumour volume cases. The infiltrative tumours had higher ctDNA than ulcero-proliferative type. Only one tumour was myoepithelial cancer on final histopathology, rest all were Squamous Cell Carcinoma (SCC). So relation of histologic type and ctDNA levels could not be commented upon. Nineteen cases of Grade I and 6 of Grade II had median pre-op ctDNA of 185.6 and 227.90, respectively (p-value 0.8987). The levels of pre-operative as well as post-operative ctDNA did not vary widely with DOI [Table/Fig-2].
Relationship of Depth of invasion with cfDNA.
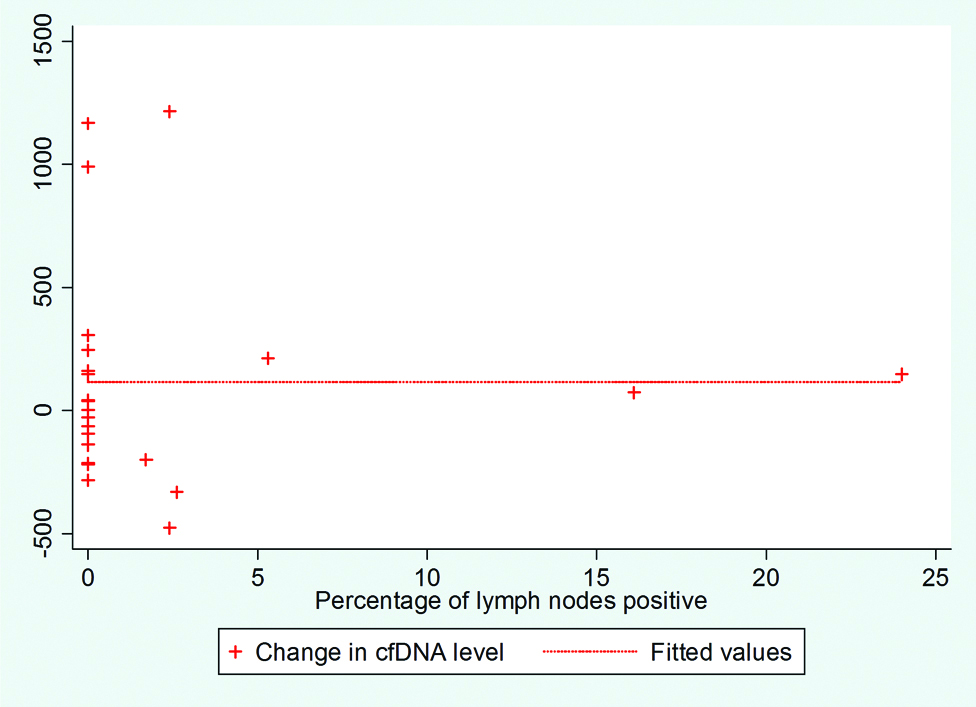
The level of pre-operative as well as post-operative ctDNA did not vary widely with margin status. Patients with clear margins had decreasing ctDNA status with treatment as compared to the close margins that had increasing trends (Median Differences between changes in ctDNA: 41.91 and -28.36, respectively, p-value: 0.8241). The ctDNA increased with increase in mitotic index, lymphocytic infiltrate, tumour budding and decreased with the amount of necrosis, and dysplasia but none reached statistical significance. The median differences for changes in ctDNA were: 17.85 for the presence of LVI and 187.27 for the absence (p-value: 0.2481). The median difference for changes in ctDNA was -7.35 for the presence of PNI and 39.325 for the absence (p-value: 0.8240). In 13 patients nodes were present and absent in rest. It was shown that ctDNA values and their differences do not correlate with the percentage of number of positive nodes among the total number of nodes dissected [Table/Fig-3].
Percentage of positive lymph nodes (among node positive cases) correlation with ctDNA.
Extra nodal extension was present in 3 out of 13 cases with positive nodes. The median difference between changes in ctDNA were 72.83 and 4.19, respectively, p-value: 0.7150. Out of 25 cases, 4 had Stage I with median pre-operative ctDNA level of 632.545 and median differences between changes in ctDNA level of 74.88; 13 had Stage II with median pre-operative ctDNA level of 160.97 and median differences between changes in ctDNA level of -34.15; 5 had Stage III with median pre-operative ctDNA level of 56.9 and median differences between changes in ctDNA level of -41.91; and 3 had Stage IVA with median pre-operative ctDNA level of 294.79 and median differences between changes in ctDNA level of 147.27. The cases with local recurrence or death had higher pre-operative ctDNA values (Mean pre-operative ctDNA were 294.725 and 185.6, respectively, p-value 0.3673).
Discussion
The incidence and mortality of oral cancer are twice as high in men (2.3% and 1.7%, respectively) compared to women (1.2% and 0.8%, respectively) [17]. Worldwide, 145,000 deaths its occur due to oral cancer and the annual incidence of diagnosed cases are 300,000. It is a serious issue especially in less developed regions [18].
In cancer patients, ctDNA forms a larger fraction of cfDNA. CtDNA is referred to as “liquid biopsy”, carrying information of tumour-related genetic and epigenetic change. A prominent clinical application of cfDNA is to monitor minimal residual disease in patients with no signs of overt metastasis and identify those individuals at increased risk for recurrence as additional treatments may aid these patients.
In the current study, authors analysed ctDNA levels pre-operatively and post-recovery from surgery. The concentration of ctDNA was calculated using qPCR, performed on CF X96 Real-Time PCR system.
The present results have shown that ctDNA levels are elevated in oral cavity cancers. In the study by Chan KCA et al., out of 105 nasopharyngeal carcinoma patients, 70% had reduction plasma ctDNA integrity index (median) after surgery from 0.356 before treatment to 0.271 at 6 weeks after treatment [19]. Present study also showed reduction in the levels. This finding follows that of Jiang WW et al., showing non-significant changes with treatment [20]. Persistent elevation of ctDNA post-recovery may be related to inflammation associated with post-operative healing or post-operative radiation therapy. Desai A et al., did not find any association of amount and length of ctDNA and DNA integrity with tumour size and histology [21].
In the present study, the mean levels of ctDNA were found to be higher among the patient with age ≤40 than that of elders. The present authors observed higher values among patients who presented with shorter duration (≤3 months) of illness than that of longer duration. Regarding personnel habits, the present authors found interesting trend of lower ctDNA levels among smokers and tobacco chewers.
It is found in this study that small sized tumour may have more ctDNA levels. However, this contradicts the previous studies showing more release of ctDNA among tumours with necrosis which is generally higher among larger tumours. Considering tumour gross appearance, the infiltrative variety which is prognostically poorer had higher levels of ctDNA than that of proliferative types. Changes in ctDNA with treatment may vary with the microscopic and macroscopic extent of the tumour. The present authors had 3 cases of skin invasion showing median difference of ctDNA (pre-operatively-post-recovery) of 213.66. This decreasing trend may indicate good response. On the other hand, the 19 tumours with muscle invasion had increasing trend. This finding does not correlate with any of the previous studies and we are unable to explain this increasing trend. The possible explanation could be aggressive disease as correlated with the smaller size of the tumour and the more limited resection of the disease for early stage.
In this study, Grade 2 has higher ctDNA denoting aggressiveness. Correlating with finding of the present study, muscle invasion and the smaller tumour, Grade I tumours had increasing trend of ctDNA with treatment as compared to the Grade II. The present authors do not find any correlation between ctDNA and depth of invasion with regards to ctDNA changes as well as pre-operative ctDNA level. This is much unexpected finding that contradicts the recent consideration of depth of invasion as an important prognostic and staging measure by AJCC 8th edition [22]. Both mitosis and necrosis are poor prognostic factors for head and neck cancer. In this study, mitosis were associated with more ctDNA. Twenty two cases had few or occasional mitosis and 3 cases had mitosis >10/hpf. However, the presence of necrosis had lower values of ctDNA than absence of necrosis. The median differences between pre-operative and post-recovery ctDNA for both LVI and PNI had shown higher values of ctDNA in this study. Five cases had LVI and four cases had PNI. The median differences for changes in ctDNA were -17.85 for the presence of LVI and 187.27 for the absence. Similarly, the median differences for changes in ctDNA were -7.35 for the presence of PNI and 39.325 for the absence treatment. In 13 patients, nodes were present and absent in 12 cases.
It was shown that ctDNA values and their differences do not correlate with the percentage of number of positive nodes among the total number of nodes dissected. This finding contradicts the study by Mazurek AM et al., found that the level of ctDNA in cases of clinical N2-N3 disease (9.28±6.34 ng/mL) was higher than in cases with clinical N0-N1 disease (7.50±3.69 ng/mL; p=0.015) [23]. Lin LH et al., also found that ctDNA levels were higher in neck lymph node positive cases as compared to negative cases [24]. Extra-nodal extension was present in 3 cases out of 13 cases with positive nodes. An inference could be made, that the positive nodes with ENE had more release of ctDNA that decreased more after the removal of such nodes as compared to the positive nodes without ENE. Lin LH et al., found that ctDNA was associated with age, gender, perineural invasion and cell differentiation. However, it was related to tumour size, TNM staging, and lympho-vascular invasion [24]. Nodes positivity denotes poor prognosis in head and neck cancers. In this study, nodes positivity does not correlate adequately with ctDNA levels and it changes with 294.79 and median differences between changes in ctDNA level of 147.27. The p-values (calculated using Kruskal Wallis equality of populations rank test) were non-significant comparing the ctDNA levels and its changes after treatment with stages. This may be due to small sample size. The only inference which could be made is that the Stage I had higher values of ctDNA as compared to the higher stage. The significance of this finding is not known. On the contrary to this, Mazurek AM et al., found higher levels in patients with Stage IV (9.16±6.04 ng/mL) compared with Stages I-III of cancer (7.26±3.63 ng/mL) (p=0.011) [23]. Desai A et al., measured statistically significant correlation of total ctDNA with nodal metastasis (p=0.001) and clinical stages (p=0.006) [21]. The current study correlates some of the findings of Coulet F et al., [25]. They found ctDNA in plasma at a higher prevalence (2 of 11 cases) when using p53 mutant allele-specific amplification. Their ctDNA did not significantly correlate with gender, tumour stage or tumour localisation significantly [25].
Finally, only two of the present cases had local recurrence and one death. None had distant recurrence. As clinicians have only one death and the average follow-up is short, overall survival cannot be calculated at this point of time. This may be better calculated if the patients will be followed longer. Similarly, disease free or progression free survival cannot be adequately commented currently. The only inference that could be made currently that the cases with local recurrence or death had higher pre-operative ctDNA values (Mean pre-operative ctDNA were 294.725 and 185.6, respectively, p-value 0.3673). Shukla D et al., concluded that progression is not correlated with changes in levels of ctDNA [26]. Lin LH et al., found that the ctDNA levels were associated with worse disease-specific survival (p=0.001) and disease-free survival (p=0.003) [24].
Thus, ctDNA is found to have important clinico-pathological and prognostic value.
Limitation(s)
Due to limited availability of financial support, the sample size remained smaller and the follow-up time was also short as the study was time bound.
Conclusion(s)
The ctDNA levels are persistently elevated in oral cavity cancers and surgical treatment of resectable oral cavity cancers may lead to the decrease of ctDNA levels. Moreover, systemic treatment like neo-adjuvant chemotherapy may have more effect on the decrease of ctDNA level. Younger (≤40 years) females with smaller size and well differentiated tumour and short (≤3 months) duration of presentation may have higher ctDNA levels. Thus, higher post-recovery ctDNA may be a marker of completeness of resection. ctDNA may be a measure of aggressiveness of tumour. A large scale prospective clinical study is required to further and better assess the predictive value of plasma ctDNA for the detection, diagnosis, treatment and prognosis of OSCC in the population.