Contamination of LJ culture media is one of the most frequent problems encountered during M. tuberculosis culture. The contaminants identified are either bacterial or fungal in origin which [1,2], suppress the growth of M. tuberculosis and thus subsequently its detection. This problem is of a more significant concern in Extrapulmonary (EP) samples which have very fewer bacilli loads. Sometimes specific contaminants behave like breakthrough contaminants [3] by completely inhibiting the growth of any acid-fast organism. In whatever case, contamination reduces the proportion of interpretable results. Such cultures are required to be repeated at an additional cost to public health systems, which causes a delay or may finally give out to diagnose tuberculosis. The ideal decontamination method should remove all microorganisms except Mycobacterium.
National Institute of Tuberculosis and Respiratory Diseases (NITRD), New Delhi is a tertiary tuberculosis care hospital in India. It has a microbiology laboratory which works as a national reference laboratory under three-tier laboratory network of RNTCP, India (Revised National Tuberculosis Control Programme) for diagnosis of tuberculosis including drug-resistant cases. Every year it receives about 20,000 clinical samples (pulmonary and EP) for identification, culture and drug susceptibility testing. Sample processing and testing are done according to steps laid in Mycobacteriology Laboratory Manual of WHO [4] which involves decontamination by N- Acetyl L- Cysteine/Sodium Hydroxide (NALC/NaOH) method followed by inoculation on LJ media which contains Malachite Green as the only antimicrobial. EP samples involve additional processing steps of grinding and cutting. Certain characteristic features are used for the preliminary identification of typical M. tuberculosis bacilli. These bacilli usually require 2-4 weeks to give visible growth from smear-positive specimens as they do not grow in primary culture in less than one week. The colonies are buff coloured (i.e., they are not pigmented), rough, and have an appearance of breadcrumbs or cauliflower. Colonies are hard/tough when touched with loop, and cannot be emulsified to smooth suspensions in saline and instead give a granular suspension. The presence of AFB is confirmed by ZN staining in which M. tuberculosis bacilli are arranged in serpentine cords of varying length. If no AFB is seen in the smear, then the growth is discarded as a contaminant.
Although in complicated tuberculosis cases, pathogenic fungi and bacteria may be present as significant co-infecting pathogens, yet their growth on LJ media in the presence of Malachite Green should always be considered as contamination. Current knowledge and data regarding a variety of such contaminants growing in LJ media are minimal. Even though few studies have been done for pulmonary samples (sputum, ET aspirate and Broncho-Alveolar lavage) [1,2,5] but none on EP samples. These studies have majorly identified bacterial and fungal organisms as the contaminants. Development of newer processing methods or improvisation of current methods will remain incomplete unless we have good knowledge about contaminant profiles from pulmonary and EP samples.
Therefore, the present study was conceptualised in NITRD hospital to add to the existing knowledge by isolating and identifying all the bacterial and fungal contaminants growing on LJ culture media in both pulmonary and EP samples.
Materials and Methods
Sample collection and processing: An observational study was carried out in the Department of Microbiology, NITRD, New Delhi, India, over the period of five months from January 2019 to May 2019 after taking institutional ethical approval (NITRD/RC/17/11253). During this period, around 6963 samples were received in the microbiology laboratory from NITRD hospital for M. tuberculosis culture. Of these, 3919 were sputum and 3044 were EP samples. No consent was ever taken from any patient because the study was done on discarded/contaminated LJ bottles. Before processing, all the sputum samples (pulmonary) were graded by Bartlett’s scoring method [6] by looking for the presence of neutrophils and squamous epithelial cells. Following the above criteria, all sputum specimens with score zero and below were not processed and regarded as rejected specimens; and remaining accepted for further processing. In this way, 2030 sputum samples were found acceptable. All the accepted sputum samples were stained by ZN stain on the same day to look for the presence of acid-fast bacilli; and then processed, irrespective of age and sex, by the NALC/NaOH method [4] and inoculated on the LJ media. All EP samples, 3044 in number, were instead directly processed by the NALC/NaOH method and then stained by ZN stain. Two LJ slopes per specimen were inoculated with 3 mm loop full of the centrifuged sediment. Cultures were incubated at 37°C until growth was observed or discarded as negative after eight weeks. Every culture was initially examined after 48-72 hours of inoculation to identify gross contamination, and after that, once in a week for up to eight weeks. Sample processing, handling and reporting were done by professionals trained for M. tuberculosis culture under biosafety cabinets strictly according to Culture and Drug Susceptibility Testing guidelines (C and DST) published by Central Tuberculosis Division, India [7].
Contaminant Identification
Bottles with any kind of growth were further processed according to gross inspection, staining and biochemical reactions [7]:
LJ slants with non-mouldy growth (could be bacteria of Mycobacterial or Non-Mycobacterial/Yeast origin) were observed for the rate of growth, gross appearance and saline suspension. Simultaneously, ZN staining was also done for these growths to look for acid fastness and thus to differentiate the organism of suspected Mycobacterial origin from those of Non-Mycobacterial/Yeast origin. Suspected mycobacterial origin colonies were sent to the main mycobacteriology section for further routine processing.
Colonies which were suspected to be of Non-Mycobacterial/ Yeast origin were now called contaminants. These were stained by Gram stain and then further processed as follows:
a) Smears which showed distinct morphology (cocci, bacilli, coccobacilli) on Gram staining were inoculated on routine bacteriology media like Chocolate Agar (CA), Blood Agar (BA) and Mackonkey Agar (MA). Any growth on these media was identified by routine biochemical methods like Catalase, Oxidase, Indole Production, Urease Production, Citrate utilisation, Triple Sugar Iron (TSI), Methyl Red Test and Voges-Proskauer Test [8]. Keeping in view the resource constraints, only those growths which could not be identified by conventional methods were subjected to MicroScan Walkaway Plus identification system for bacteria [9].
b) Growths showing Budding Yeast Cells (BYC) were proposed to be inoculated onto SDA for further culture.
Lowenstein Jensen slant with Cottony/Mouldy colonies in both bottles was presumed to be contaminated with fungal organisms and inoculated on Sabouraud’s Dextrose Agar (SDA). Subsequent growth on SDA slants was identified based on gross morphology, Lacto Phenol Cotton Blue (LPCB) preparation and slide culture techniques [10].
A sample was labelled as contaminated only when both the LJ slants of the pair was found to be contaminated by the organisms of non-mycobacterial/yeast origin [7]. Calibrated thermometers and biological indicators were routinely used to check the incubator and autoclave performance [11].
Statistical Analysis
Statistical analysis was done using SPSS version 20 (IBM®, New York, NY, USA). Chi-square test was performed. A p-value of <0.05 was considered indicative of a statistically significant difference.
Results
A total of 5074 LJ pairs (10148 bottles), irrespective of age and sex, were inoculated during the five-month study period [Table/Fig-1] out of which 40% (2030/5074) were inoculated with pulmonary samples and 60% (3044/5074) with EP samples. A total of 2.2% (112/5074) LJ pairs (224 bottles) were found to be contaminated, taking overall CR in LJ media to 2.2%. Individually, CR was 2.9% (60/2030) in pulmonary samples and 1.7% (52/3044) in EP samples (p<0.05). In addition to these pairs, there were 1.5% (77/5074) LJ pairs where only one of the pair bottles was contaminated. Therefore, these 77 pairs were not accounted for in computing the overall CR. Nevertheless, overall, there were 301 (112+112+77) bottles out of 10148, which grew contaminants during the study.
Distribution of contaminated and non-contaminated LJ slant pairs. (n=5074).
| LJ slant pairs inoculated with samples |
|---|
| Pulmonary | Extra pulmonary | Total |
|---|
| Contaminated | 60 | 52 | 112 |
| Non contaminated | 1970 | 2992 | 4962 |
| Total | 2030 | 3044 | 5074 |
Amongst these 301 contaminated LJ bottles, bacterial and fungal contaminants were found in 87.7% (264/301) and 12.3% (37/301) bottles, respectively. No bottle had a mixture of bacterial and fungal growth. A massive 78% (206/264) of the bacterially contaminated bottles were associated with a change in the physical appearance of the media (media liquefaction/breaking/crumbling). No change in media appearance was seen in fungal contaminated bottles [Table/Fig-2].
Growth of Mycobacterium tuberculosis, bacterial contaminants and fungal contaminants on LJ Media.
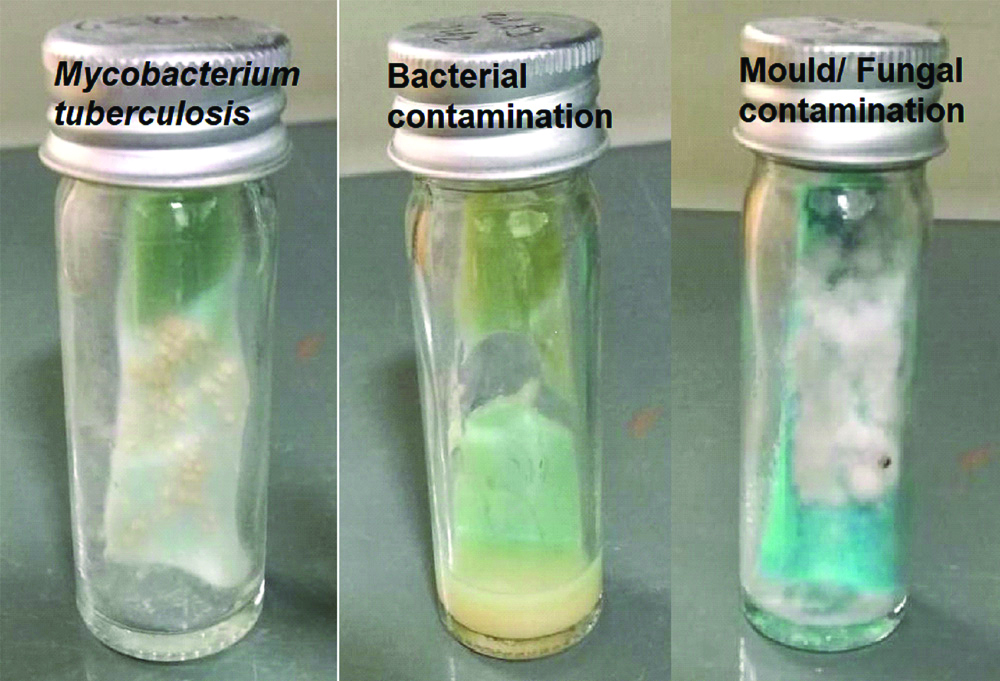
Amongst 264 bacterial contaminated bottles, 262 bottles had an only single type of bacterial organism. Only two EP (pleural pus) bottles had a mixture of two organisms (2×2=4). Therefore a total of 266 (262+4) different bacterial organisms were identified. On the contrary, out of 37 fungal contaminated bottles, no bottle had a mixture of fungal organisms. Therefore, overall 303 organisms (266+37) were isolated, either singly or as a mixture, from 301 contaminated bottles, amongst which 87.8% (266/303) were bacteria and 12.2% (37/303) fungal. Bacterial contaminants were found more in pulmonary samples, whereas fungal were found more in EP samples (p<0.05).
In the study, 12 different genera of bacterial contaminants were grown [Table/Fig-3]. At 42.6% (129/303), Aerobic Spore Bearers (Bacillus spp) were the most frequent bacterial isolates, followed by 23.1% (70/303) Pseudomonas isolates. The two pleural pus bottles, which had mixed bacterial growth, were found to have Pseudomonas and Staphylococcus sciuri. Interestingly, 19 bacterial contaminants could not be cultured on any of the routine bacteriology media; although on direct Gram staining of LJ slant growth, they did show distinct morphology (cocci, bacilli or coccobacilli).
Bacterial contaminants isolated from LJ slants (either singly or as mixture) (n=303).
| Organisms | LJ pulmonary | LJ extrapulmonary | Total (%) n=303 | p-value | Total |
|---|
| Gram positives | ASB | 87 | 42 | 129 | <0.05 | <0.05 | =141/303$ (46.5%) |
| CONS | 4 | 5 | 9 | 0.554 |
| -S.epidermidis | 3 | 2 | 5 | 0.790 |
| -S.haemolyticus | 1 | 1 | 2 | 0.903 |
| -S. sciuri* | 0 | 2* | 2* | # |
| Micrococcus | 0 | 1 | 1 | # |
| Enterococcus faecalis | 0 | 1 | 1 | # |
| Listeria | 1 | 0 | 1 | # |
| Gram negative | Pseudomonas@ | 35 | 35@ | 70@ | 0.429 | 0.106 | =106/303$ (35%) |
| Escherichia coli | 6 | 8 | 14 | 0.632 |
| Citrobacter | 3 | 0 | 3 | # |
| Klebsiella pneumoniae | 0 | 4 | 4 | # |
| Serratia | 2 | 4 | 6 | 1.065 |
| Proteus mirabilis | 5 | 1 | 6 | 2.103 |
| Acinetobacter baumannii | 0 | 3 | 3 | # |
| No growth on CA, BA and MAC | 11 | 8 | 19 | 0.733 | =19/303 (6.3%) |
| Total | 154 | 112 | 266 | <0.05 | =266/303 (87.8%) |
*2 Staphylococcus sciuri isolates grew along with Pseudomonas as a mixture in EP bottles (pleural fluid).
@2 Pseudomonas isolates grew along with Staphylococcus sciuri as a mixture in EP bottles (pleural pus)
ASB: Aerobic spore bearers; CONS: Coagulase negative Staphylococcus spp (S. epidermidis, S. haemolyticus, S. sciuri)
#Significance could not be calculated because of the small size
$303 includes organisms isolated as mixed growth
All the 37 fungal isolates grown were moulds amongst which 22 were of genus Aspergillus (A.flavus, A.fumigatus and A.niger) [Table/Fig-4]. Some of the other moulds identified were Penicillium, Scopulariopsis and Alternaria. Eight moulds could not be identified even after repeated attempts by conventional techniques like gross morphology, LPCB preparation and slide culture techniques [10]. These were white, non-sporulating in nature and consisted of long parallel bundles of thin septate hyphae and were present in higher amounts in EP samples (p<0.05). Interestingly, no BYC/Candida was ever identified during the study.
Fungal contaminants isolated from LJ Media.
| Organisms | LJ pulmonary | LJ extra pulmonary | Total | p-value | Total |
|---|
| A flavus | 4 | 8 | 12 | 0.140 | 22/303 (7.3%) |
| A. fumigatus | 2 | 5 | 7 | 1.885 |
| A.niger | 1 | 2 | 3 | 0.467 |
| Unidentified white mould | 1 | 7 | 8 | <0.05 | 8/303 (2.6%) |
| Penicillium | 2 | 3 | 5 | 0.452 | 6/303 (2%) |
| Scopulariopsis | 0 | 1 | 1 | # |
| Alternaria | 0 | 1 | 1 | # | 1/303 (0.3%) |
| Total | 10 | 27 | 37 | <0.05 | 37/303 (12.2%) |
#Significance could not be calculated because of a very small size
Out of 60 contaminated pulmonary bottle pairs, as isolated above, 15% (9/60) were positive on ZN smear microscopy (done before inoculating on LJ media). On the other hand, amongst 52 contaminated EP bottle pairs, only 3.6% (2/56) were ZN smear-positive. Amongst 77 LJ pairs, which had only one of the bottles infected, none was ZN smear-positive. Therefore, overall ZN smear positivity rate was 3.75% (11/301) amongst contaminanted LJ bottle pairs. Interestingly, at no point during the study, did any of the contaminated LJ bottles have mixed growth of acid-fast and non-acid fast organisms (now called contaminants). Moreover, at no point was a mixture of bacterial and fungal organisms isolated.
Discussion
Although in the era of molecular diagnostics, M. tuberculosis detection has become easy and fast, yet the importance of conventional LJ culture methods cannot be ruled out especially in the fund and resource-constrained countries. As a general rule, a CR of 2-3% is acceptable in solid LJ media during fresh specimen processing [7]. Processing delays beyond three days may increase CR to as high as 5% [7]. In the present study, where samples were collected, processed and inoculated on the same day, overall CR was 2.2%, which was well within the acceptable limits. At other centres worldwide, where studies have been done primarily on pulmonary samples, these rates were found to vary widely from 4% to 30.1% [1,2,12-15]. Even though CR of 2.2% in the present work was well within the acceptable range of 5%, efforts should be constituted to bring it down farther. A likely contributor to the observed low CR in the present study is the sample processing and media preparation by highly experienced personnel at NITRD, a national reference laboratory. The suboptimal CR in other studies could be due to factors like high sample load, space limitation in the laboratory, frequent breakdown of old instruments, electricity failure and never-ending construction work of laboratories, all of which are unique in developing countries.
Contamination is possible in any of the several ways: (1) inadequate cleaning of body site from where clinical sample is collected; (2) Improper sample handling techniques leading to sterilisation failure during collection, transport or processing; and (3) Inadequate sterilisation of instruments required during M. tuberculosis culture; (4) Inherent inadequacy of NALC/NaOH method to remove all the contaminants. It is expected that if there is any problem in sample collection, handling or sterilisation practices then the CR would be equal in both pulmonary and EP bottles. However, in the present study, it was observed that 2.9% CR of pulmonary bottles was significantly higher than 1.7% CR of EP bottles (p<0.05). This apparent inadequacy of NALC/NaOH method in removing contaminants equally from pulmonary and EP samples could be because pulmonary samples sites are already heavily loaded with commensals organisms and have environmental contaminant exposure all the time through oral and inhalation route. This point is further authenticated by isolation of ASB (Bacillus spp) in higher proportion from pulmonary samples (p<0.05), which are known to be naturally present in higher amount there [16,17].
In the current study, only 12 different genera of bacteria were isolated, the variety of which was quite different from the previously done studies [1,2,15] in which organisms like Burkholderia sp, Achromobacterium sp, Corynebacterium sp, Lactobacillus rhamnosus, Rahnella aquatilis and Sphingobacterium spiritivorum were also reported. This could be partly explained by the limitation of this study in using mainly conventional phenotypic bacterial identification methods and opting for automated methods like MicroScan Walkaway [9] only when these conventional methods failed.
Among fungal contaminants, EP bottles were found to be more contaminated than pulmonary bottles (p<0.05). This could be due to additional processing steps of grinding and cutting involved with them, which attract fungal spores from the environment. Higher fungal contaminants in EP samples are a matter of concern since these samples are difficult to obtain. Environmental contamination can be prevented by processing samples in biosafety cabinets of higher class like Class II B2/B3 or Class III. Aspergillus spp 22 (7.3%), which grew as the most common mould in this study is globally recognised as the most frequent fungal isolate from pulmonary tuberculosis cases and is also implicated in post tubercular sequelae as one of the causative organisms [18]. A significantly high number of non-sporulating moulds were found in EP samples (p<0.05). Although no high-end molecular tests were done to characterise them, these could be basidiomycetes and ascomycetes [19].
At no point during the current study did any of the contaminant grow along with acid-fast organism. Although 3.7% (11/301) bottles were positive for acid-fast organisms before culturing, their growth was suppressed in the presence of other contaminants. It indicates that probably all the contaminants act as breakthrough agents by completely inhibiting the growth of any acid-fast bacilli, if at all present in the sample. These 11 bottles were false culture negative. This observation becomes quite relevant in the context of EP samples wherein M. tuberculosis bacilli load is already very less. LJ culture is more sensitive than ZN smear microscopy for detection of M. tuberculosis growth [20]; thus, no growth of acid-fast organisms in contaminated bottles further supports the inference that such organisms completely inhibit acid-fast bacilli growth.
Furthermore, there were no LJ sets in which both the bacterial and fungal contaminants grew together. Partly be explained by the fact that many of the bacteria cultured during this study inhibit fungal growth [21-23]. The absence of budding yeast cells/ Candida could not be explained by existing literature knowledge and needs to be further explored.
Limitation(s)
MicroScan Walkaway Plus identification system could not be used on all the bacterial isolates because of resource constraints. The absence of budding yeast cells/Candida amongst various contaminants could not be explained by existing literature knowledge and needs to be further explored. High-end molecular techniques like liquid culture (MGIT), Line probe assay and Cartridge based nucleic acid amplification technique (GeneXpert) could have been used to know precisely how much TB positive cases were missed due to contamination. DNA sequencing could have been done to identify white non-Sporulating moulds.
Conclusion(s)
The present study gives insight into the kind of bacterial and fungal contaminants growing on LJ Media. Breakthrough nature of contaminants indicates that they probably act by completely inhibiting the growth of any acid-fast bacilli present in the sample. This observation becomes quite relevant in EP samples wherein M. tuberculosis bacilli load is already very less. This M. tuberculosis growth masking can thus decrease the overall sensitivity of LJ culture. In the past, such studies were biased toward pulmonary samples. The present study focuses on both pulmonary and EP samples. The presence of such breakthrough contaminants raises the possibility of missing many tuberculosis cases in developing countries- a lost opportunity.
*2 Staphylococcus sciuri isolates grew along with Pseudomonas as a mixture in EP bottles (pleural fluid).
@2 Pseudomonas isolates grew along with Staphylococcus sciuri as a mixture in EP bottles (pleural pus)
ASB: Aerobic spore bearers; CONS: Coagulase negative Staphylococcus spp (S. epidermidis, S. haemolyticus, S. sciuri)
#Significance could not be calculated because of the small size
$303 includes organisms isolated as mixed growth
#Significance could not be calculated because of a very small size