Introduction
Breast cancer is the most commonly diagnosed cancer among women world-wide. The incidence and mortality age-standardised rates are 31.3% and 14.9%, respectively in the lower Human Development Index (HDI) regions including India [1]. As per Globocan 2018, in India, the number of new breast cancer cases in 2018 is 1,62,468 (27%) among 5,87,249 new cases of all cancers in females [1]. The number of projected cases for breast cancer in India for 2020 is 1,79,790 [2]. The diverse molecular characteristics of breast cancer cause differences in prognoses, patterns of recurrence and sensitivity to therapies [3-5]. In addition to the traditional gold standard, histopathological characterisation of the primary tumour and regional lymph nodes, the latest cancer staging manual [5] has endorsed the documentation of several biomarkers at the time of initial diagnosis of breast cancer including histologic grade, Hormone Receptor (HR) status and HER2, Ki-67/mitotic count. These biomarkers influence prognosis and probability of response to systemic therapies [3,6].
Ki67 index assessment by IHC is the current assay of choice for proliferation analysis [7-10]. Assessment of Ki67 status in breast cancer has a potential role in standard clinical practice as a prognostic and predictive marker [8,11-13]. It is identified that Ki67 expression corresponds linearly with tumour progression [9,14]. The International Ki67 Working Group recommends the use of Ki67 status in the context of clinicopathological factors and IHC biomarkers (ER, PR, HER2) [13]. Studies support the view that Ki67 is continuous marker, considering the continuous disparity of the proliferation rate in different tumours depending on cohort characteristics, molecular subtype and clinical setting. High and low values of Ki67 are reproducible and clinically useful; however no “optimal” cut-point [12,15] or a standard operating procedure [5] is available and one should stop looking for it [12,15]. The International Ki67 Working Group, considered the Ki67 scoring in three categories <10%, 10-20%, and >20% and showed a very strong inter-observer agreement on cases with scores <10% and >20% than the intermediate range of 10-20% [13]. Due to inter-observer variance and intra-tumoural heterogeneity, the standardisation of intermediate levels of Ki67 is difficult; St Gallen guidelines state that intermediate proliferation rate should not be used for clinical decisions [12,16].
Tumour Protein 53 (TP53) is a tumour suppression gene that encodes p53 [17], a transcription factor implicated in the cell cycle regulation, DNA integrity, cellular aging, apoptosis, autophagy, mitotic catastrophe, and angiogenesis [18]. In the spectrum of somatic TP53 mutations identified in breast cancer in a meta-analysis [19] and also reported by TP53 mutations database [20], the most common are point mis-sense mutations [17,19,20]. The p53 IHC staining is based on the fact that the mutant protein is stabilised and thus accumulates in the nucleus of malignant cells, enabling its detection [19,21-23]. Lower IHC sensitivity (72%) and specificity (92%) to detect p53 mutations, as compared with sequencing of cDNA is attributed to the rare TP53 mutations (about 20%) leading to protein truncation and thus not identifiable by IHC [24]. As per the College of American Pathologists Consensus 1999, optimal methodology does not exist for either molecular or IHC assays [25]. Due to the time and cost involved in the sequencing of TP53 gene, IHC is the most practical and prevailing modality to detect p53 mutations [6,18].
Although gene expression profiling is commercially available to define the molecular sub-types, inaccessibility and high cost precludes its use in routine diagnostics [5,15]. IHC evaluation of ER, PR, HER2 and Ki67 status in combination with Nottingham Grading System (NGS) forms the backbone to classify breast cancers into surrogates of the genetically defined subtypes [5,26]. Many studies [9-11,14,26-29] have analysed and reported the relationships of these biomarkers with one another; whereas p53 status is not a standard parameter assessed in breast carcinoma. Few studies [30-34] have analysed p53 status along with traditional parameters, however there is paucity of literature [8,35] with respect to the analysis of combined Ki67-p53 status in breast carcinoma. The present study was conducted to analyse Ki67 and p53 status in breast carcinoma; their relationship with traditional immuno-markers and clinico-pathological characteristics. An attempt to further stratify the HR positive breast carcinomas into favourable and unfavourable phenotypes based on the combined Ki67-p53 status was made.
Materials and Methods
This was an observational study conducted at Vydehi Institute of Medical Sciences and Research Centre between August 2014 to April 2016. Ethical clearance for the study was obtained from the Institutional Ethics Committee (VIMS and RC/IEC/011/2014-15).
Inclusion criteria: All newly diagnosed female breast cancer patients of all age groups with histological diagnosis of invasive carcinoma were included for the study.
Continuous sampling method was adopted. The sample size was determined using the following formula:
n=z2p (1-p)/d2,where n=sample size.
Using z=Z score for 95% confidence level, p=population proportion of p53, 15% as reported by Hill KA et al., [36] and d=margin of error, 0.1 the sample size was calculated to be a minimum of 49. A total of 50 consecutive cases were enrolled for the study.
The clinical characteristics of these cases including age of patient, age at menarche and menopausal status were obtained from the case files.
Exclusion criteria: The cases undergoing or started with treatment and having pathological complete response or clinical partial response were not included in the study.
The surgical specimens were fixed in 10% buffered formalin, routinely processed, paraffin-embedded and the sections were stained with haematoxylin and eosin stain. The final diagnosis of the tumour histotype was made in accordance with World Health Organisation classification of breast tumours [37]. The tumours were graded according to NGS [38,39].
Immunohistochemistry
IHC evaluation of the markers ER, PR, HER2, Ki67 and p53 were done on representative histologic sections. Quality control was ensured by evaluation of appropriate known positive control and negative control run for each batch of all the markers. For ER and PR, normal breast duct epithelia when present in the sections were used as internal positive controls and uterine cervix was used as external positive control. For HER2 and p53, the positive controls used were, known case of invasive carcinoma, NST tumour with HER2 amplification and p53 overexpression respectively. Lymphoid follicles of appendix tissue were used as positive control for Ki67. The negative control sections were made by excluding the respective primary antibody. The results were assessed and scored independently by two pathologists, and cases with disparate scores were re-evaluated and discussed until a consensus was reached.
IHC staining for ER, PR, HER2 and Ki67 was carried out using Ventana automated IHC slide staining device (Ventana Medical Systems) following the manufacturer’s guidelines. The reaction was visualised by ultraView Universal DAB detection kit comprising multimer HRP (Horse-radish Peroxidase) labelled secondary antibody and DAB chromogen. Ventana rabbit monoclonal Ready To Use (RTU) primary antibodies, clone SP1, IE2 and 4B5 were used for ER, PR and HER2, respectively. Ki67 antigen was identified using mouse monoclonal anti-Ki67 antibody, RTU, clone GM001, PathnSitu. The IHC assay for p53 antigens was performed with Dako Autostainer Link 48 using mouse monoclonal anti-p53 antibody, RTU, clone DO-7, Dako. The reaction was visualised by EnVision FLEX, High pH (Link) comprising polymer HRP labelled secondary antibody.
IHC Evaluation
The ER and PR slides were scored for both intensity (0-3) and %-positivity (0-5) of nuclear staining and with a total score classified as ER+ or ER- and PR+ or PR- as per the Allred system and ASCO/CAP guideline recommendations [40]. ASCO/CAP guidelines [41] were followed to determine the HER2 status. The slides were scored on a scale of 0 to 3+ for membrane staining pattern; reflex testing with FISH (Fluorescent in-situ hybridization) was ordered for cases with 2+ (equivocal) score. For Ki67 evaluation, the entire invasive tumour area was analysed under high power (400x) and a range of 500-1000 tumour cells (depending on the cellularity) were counted manually [7,8]. Ki67 score was calculated as the percentage of total number of invasive tumour cell nuclei positively stained, divided by the total number of invasive tumour cell nuclei counted across all fields [8,13,27]. Staining for Ki67 antigen was considered positive when there was any brown stain in the tumour nuclei [7] above background and negative when the tumour nuclei showed only a blue counter-stained nucleus [13]. The Ki67 score was categorised as negative (<20%), and positive (≥20%) [42]. For interpretation of p53 IHC staining, a visual score of 10% or more nuclear stain positivity irrespective of intensity in invasive tumour cells was defined as positive [6,8,30,31,34]. Based on the expression of ER, PR, HER2 and Ki67, the cases were categorised into surrogate molecular subtypes as follows: Luminal Like (HR-positive and HER2-negative: Luminal A-like, Luminal B-like), HER2-Like (HER2 positive and HR negative or HER2 positive and HR positive) and Basal-Like (Triple negative: ER, PR, HER2 negative) [5,15]. Luminal cases were stratified into A-like and B-like depending on the Ki67-low and high values respectively. The HR-positive cases were categorised into ‘favourable’ (Ki67-low and p53-negative) and ‘unfavourable’ (Ki67-low and p53-positive, Ki67-high and p53-negative, Ki67-high and p53-positive) phenotypes [8].
Statistical Analysis
Distribution of the markers ER, PR, HER2, Ki67 and p53 expression in all cases of breast carcinoma, along with clinico-pathological parameters were expressed as percentage/proportion using descriptive statistics. Association between IHC expression of Ki67 and p53 and the status of the IHC markers ER, PR, HER2 and various clinico-pathological parameters such as age, menopausal status, tumour size, histologic grade, nodal status, were correlated using Chi-square test or Fisher’s-exact test. The p-value was calculated to ascertain a statistical significance. A p-value of <0.05 was considered statistically significant.
Results
The distribution of clinico-pathological characteristics of the 50 patients included in this study are summarised in [Table/Fig-1]. Representative images of IHC staining for the markers analysed in this study are shown in [Table/Fig-2,3 and 4]. In 7 cases, the IHC HER2 testing result was 2+ (equivocal); for these cases reflex testing with FISH was ordered on the same specimen and two turned out positive. Of the 50 cases, 33 were HR positive (ER/PR positive) including 23 Luminal-like and 10 HER2-like. Depending on Ki67 value, the 23 Luminal type were further subtyped as 17 Luminal-A like (Ki<20%) and 6 Luminal-B like (Ki≥20%) [Table/Fig-5]. Statistically significant association among molecular subtypes of cases with age, menopausal status and histologic grade was noted [Table/Fig-6]. The correlation of Ki67 status and p53 status with clinico-pathological characteristics and IHC based molecular subtypes are shown in [Table/Fig-7,8 and 9]. Among 33 HR-positive tumours, there were 14 (42%) Ki67-high tumours and 15 (45.5%) p53-positive tumours.
Clinicopathological characteristics of the cases (n=50).
| Characteristics | Pre-menopausal n=23 (46%) | Post-menopausal n=27 (54%) | Number of cases n=50 (%) |
|---|
| Median age, years (Range) | 55 (30-70) |
| Histological type | | | |
| Invasive carcinoma, NST | 23 (46) | 25 (50) | 48 (96) |
| Mucinous carcinoma | 0 (00) | 2 (04) | 2 (04) |
| Tumour grade | | | |
| G1 | 3 (06) | 3 (06) | 6 (12) |
| G2 | 15 (30) | 19 (38) | 34 (68) |
| G3 | 5 (10) | 5 (10) | 10 (20) |
| pT stage (cm) | | | |
| T1 (≤2) | 3 (06) | 2 (04) | 5 (10) |
| T2 (>2 ≤5) | 9 (18) | 15 (30) | 24 (48) |
| T3 (>5) | 10 (20) | 9 (18) | 19 (38) |
| T4 (any size*) | 1 (02) | 1 (02) | 2 (04) |
| Nodal status | | | |
| Negative | 9 (18) | 10 (20) | 19 (38) |
| Positive | 14 (28) | 17 (34) | 31 (62) |
| Lympho-vascular invasion | | | |
| Absent | 7 (14) | 5 (10) | 12 (24) |
| Present | 16 (32) | 22 (44) | 38 (76) |
*Tumour any size with direct extension to the chest wall and/or to the skin
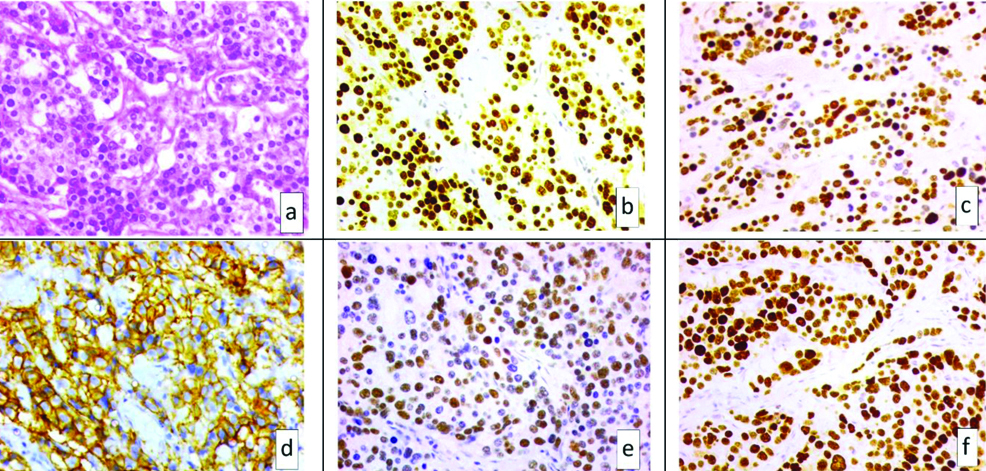
a) H&E microphotograph of a case of invasive carcinoma, NST (x400) showing; b) positive ER staining (x400); c) positive PR staining (x400); d) positive Her2 staining (x400); e) positive Ki 67 staining (x400) and f) positive p53 staining (x400).
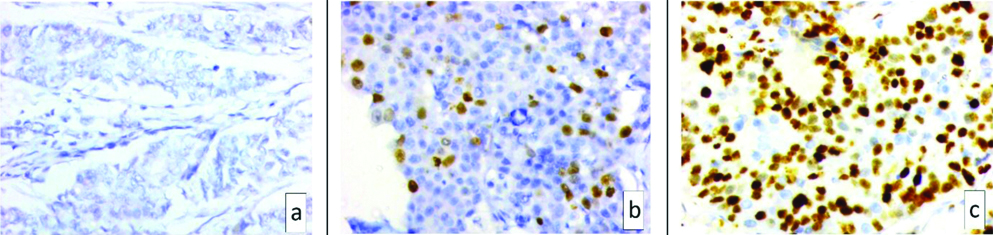
Microphotograph of Ki 67 showing a) <10% positive staining (x400); b) 10-20% positive staining (x400) and c) >20% positive staining (x400).
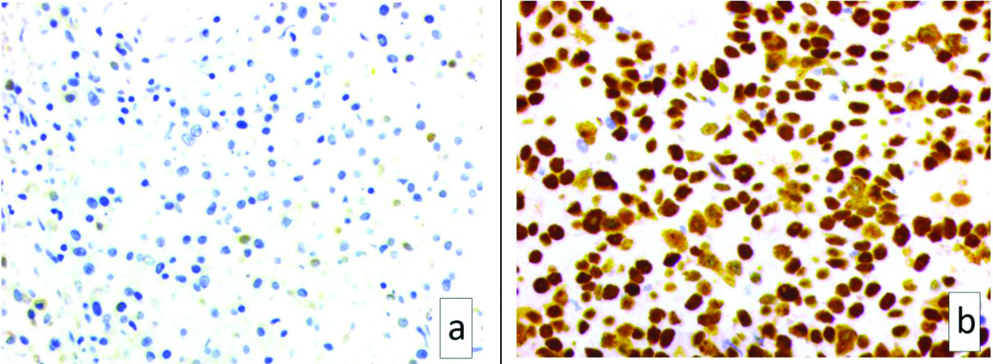
Microphotograph of p53 showing a) negative (<10%) staining (x400) and b) positive (>10%) staining (x400).
Expression of IHC markers and clinically defined-therapy oriented subtypes of breast cancer cases (n=50).
| Expression of markers | Pre-menopausal n=23 (46%) | Post-menopausal n=27 (54%) | Number of cases n=50 (%) |
|---|
| ER | | | |
| Negative | 7 (14) | 11 (22) | 18 (36) |
| Positive | 16 (32) | 16 (32) | 32 (64) |
| PR | | | |
| Negative | 13 (26) | 13 (26) | 26 (52) |
| Positive | 10 (20) | 14 (28) | 24 (48) |
| HER 2 | | | |
| Negative | 11 (22) | 24 (48) | 35 (70) |
| Positive | 12 (24) | 3 (06) | 15 (30) |
| Ki67 | | | |
| Low (<20%) | 11 (22) | 14 (28) | 25 (50) |
| High (≥20%) | 12 (24) | 13 (26) | 25 (50) |
| p53 | | | |
| Negative | 8 (16) | 18 (36) | 26 (52) |
| Positive | 15 (30) | 9 (18) | 24 (48) |
| Molecular subtypes | | | |
| Luminal A like (HR + HER2-, low Ki67) | 6 (12) | 11 (22) | 17 (34) |
| Luminal B like (HR + HER2-, high Ki67) | 2 (04) | 4 (08) | 6 (12) |
| HER2 like | | | |
| HER2+, HR+ | 8 (16) | 2 (04) | 10 (20) |
| HER2+, HR- | 4 (08) | 1 (02) | 5 (10) |
| Basal like (ER-, PR-, HER2-) | 3 (06) | 9 (18) | 12 (24) |
HR: Hormone receptor; +: Positive; -: Negative
Correlation of clinicopathological features with clinically defined-therapy oriented subtypes of breast cancer cases (n=50).
| Characteristics | Luminal A like (n=17) | Luminal B like (n=6) | HER2 like (n=15) | Basal like (n=12) | p-value† |
|---|
| | HER2+ HR+ (n=10) | HER2+ HR- (n=5) | |
|---|
| Age (years) | | | | | | |
| <50 (n=20) | 4 | 1 | 8 | 4 | 3 | 0.006 |
| >50 (n=30) | 13 | 5 | 2 | 1 | 9 |
| Menopausal status | | | | | | |
| Premenopausal (n=23) | 6 | 2 | 8 | 4 | 3 | 0.036 |
| Postmenopausal (n=27) | 11 | 4 | 2 | 1 | 9 |
| Histological type | | | | | | |
| Invasive carcinoma, NST (n=48) | 17 | 5 | 10 | 5 | 11 | 0.33 |
| Mucinous carcinoma (n=2) | 0 | 1 | 0 | 0 | 1 |
| Tumour grade | | | | | | |
| G1 (n=6) | 4 | 0 | 1 | 0 | 1 | 0.007 |
| G2 (n=34) | 13 | 1 | 7 | 4 | 9 |
| G3 (n=10) | 0 | 5 | 2 | 1 | 2 |
| pT stage (cm) | | | | | | |
| T1 (≤2) (n=5) | 3 | 0 | 1 | 1 | 0 | 0.46 |
| T2 (>2 ≤5) (n=24) | 9 | 2 | 5 | 1 | 7 |
| T3 (>5) (n=19) | 5 | 3 | 4 | 2 | 5 |
| T4 (any size*) (n=2) | 0 | 1 | 0 | 1 | 0 |
| Nodal status | | | | | | |
| Negative (n=19) | 10 | 2 | 5 | 0 | 2 | 0.056 |
| Positive (n=31) | 7 | 4 | 5 | 5 | 10 |
| Lympho-vascular invasion | | | | | | |
| Absent (n=12) | 6 | 1 | 4 | 0 | 1 | 0.227 |
| Present (n=38) | 11 | 5 | 6 | 5 | 11 |
*Tumour any size with direct extension to the chest wall and/or to the skin
†Calculated using Fishers exact Test
Correlation of Ki67 and p53 expression with clinicopathological features of breast cancer cases (n=50).
| Characteristics | Ki67 | p-value | p53 | p-value |
|---|
| Low (n=25) | High (n=25) | Negative (n=26) | Positive (n=24) |
|---|
| Age (Years) | | | | | | |
| <50 (n=20) | 9 | 11 | 0.77** | 6 | 14 | 0.024** |
| >50 (n=30) | 16 | 14 | 20 | 10 |
| Menopausal status | | | | | | |
| Premenopausal (n=23) | 11 | 12 | 0.776** | 8 | 15 | 0.049** |
| Postmenopausal (n=27) | 14 | 13 | 18 | 9 |
| Histological type | | | | | | |
| Invasive carcinoma, NST (n=48) | 24 | 24 | 1.0** | 24 | 24 | 0.5** |
| Mucinous carcinoma (n=2) | 1 | 1 | 2 | 0 |
| Tumour grade | | | | | | |
| G1 (n=6) | 6 | 0 | 0.00006† | 5 | 1 | 0.36† |
| G2 (n=34) | 19 | 15 | 17 | 17 |
| G3 (n=10) | 0 | 10 | 4 | 6 |
| pT stage (cm) | | | | | | |
| T1 (≤2) (n=5) | 3 | 2 | 0.7† | 2 | 3 | 0.47† |
| T2 (>2 ≤5) (n=24) | 13 | 11 | 14 | 10 |
| T3 (>5) (n=19) | 9 | 10 | 10 | 9 |
| T4 (any size*) (n=2) | 0 | 2 | 0 | 2 |
| Nodal status | | | | | | |
| Negative (n=19) | 13 | 6 | 0.08** | 15 | 4 | 0.007** |
| Positive (n=31) | 12 | 19 | 11 | 20 |
| Lympho-vascular invasion | | | | | | |
| Absent (n=12) | 7 | 5 | 0.7** | 6 | 6 | 0.8** |
| Present (n=38) | 18 | 20 | 20 | 18 |
*Tumour any size with direct extension to the chest wall and/or to the skin
†Calculated using Fishers exact Test; **Calculated using Chi square test with Yates’ correlation
Correlation of Ki67 expression with ER, PR, HER2 status and subtypes of breast cancer cases (n=50).
| Characteristics | Ki67 | p-value |
|---|
| Low (n=25) | High (n=25) |
|---|
| ER | | | |
| Negative (n=18) | 7 | 11 | 0.37* |
| Positive (n=32) | 18 | 14 |
| PR | | | |
| Negative (n=26) | 12 | 14 | 0.77* |
| Positive (n=24) | 13 | 11 |
| HER2 | | | |
| Negative (n=35) | 22 | 13 | 0.013* |
| Positive (n=15) | 3 | 12 |
| Molecular subtypes | | | |
| Luminal like (n=23) | 17 | 6 | 0.009† |
| HER2 like | | |
| HER2+, HR + (n=10) | 2 | 8 |
| HER2+, HR- (n=5) | 1 | 4 |
| Basal like (n=12) | 5 | 7 |
*Calculated using Chi square test with Yates’ correlation
†Calculated using Fishers exact Test
HR: Hormone receptor; +: Positive; -: Negative
Correlation of p53 expression with ER, PR, HER2, Ki67 status and subtypes of breast cancer cases (n=50).
| Characteristics | p53 | p-value |
|---|
| Negative (n=26) | Positive (n=24) |
|---|
| ER | | | |
| Negative (n=18) | 8 | 10 | 0.6* |
| Positive (n=32) | 18 | 14 |
| PR | | | |
| Negative (n=26) | 10 | 16 | 0.08* |
| Positive (n=24) | 16 | 8 |
| HER2 | | | |
| Negative (n=35) | 20 | 15 | 0.42* |
| Positive (n=15) | 6 | 9 |
| Molecular subtypes | | | |
| Luminal A like (n=17) | 10 | 7 | 0.67† |
| Luminal B like (n=6) | 3 | 3 |
| HER2 like | | |
| HER2+, HR +(n=10) | 5 | 5 |
| HER2+, HR- (n=5) | 1 | 4 |
| Basal like (n=12) | 7 | 5 |
| Ki67 | | | |
| Low (<20%) (n=25) | 15 | 10 | 0.39* |
| High (≥20%) (n=25) | 11 | 14 |
*Calculated using Chi square test with Yates’ correlation
†Calculated using Fishers exact Test
HR: Hormone receptor; +: Positive; -: Negative
The cases with “favourable” Ki67-low and p53-negative phenotype (n=12) were predominantly post-menopausal and these tumours showed lower frequencies of high nuclear grade and nodal involvement than did those with “unfavourable” phenotype. Majority of the HER2-positive tumours were unfavourable phenotype tumours [Table/Fig-10].
Correlation of combined Ki67 –p53 status with clinicopathological features in HR-positive breast cancer cases (n=33).
| Characteristics | Combined Ki67-p53 status | p-value† |
|---|
| Favourable phenotype | Unfavourable phenotype |
|---|
| Ki67 Low p53 Negative (n=12) | Ki67 Low p53 Positive (n=7) | Ki67 High p53 Negative (n=6) | Ki67 High p53 Positive (n=8) |
|---|
| Menopausal status | | | | | |
| Premenopausal (n=16) | 4 | 4 | 1 | 7 | 0.037 |
| Postmenopausal (n=17) | 8 | 3 | 5 | 1 |
| Tumour grade | | | | | |
| G1 (n=5) | 4 | 1 | 0 | 0 | 0.001 |
| G2 (n=20) | 8 | 6 | 4 | 2 |
| G3 (n=8) | 0 | 0 | 2 | 6 |
| pT stage (cm) | | | | | |
| T1 (≤2) (n=4) | 2 | 1 | 0 | 1 | 0.753 |
| T2 (>2 ≤5) (n=16) | 6 | 4 | 4 | 2 |
| T3 (>5) (n=13) | 4 | 2 | 2 | 5 |
| T4 (any size*) (n=0) | 0 | 0 | 0 | 0 |
| Nodal status | | | | | |
| Negative (n=17) | 9 | 3 | 4 | 1 | 0.04 |
| Positive (n=16) | 3 | 4 | 2 | 7 |
| Lympho-vascular invasion | | | | | |
| Absent (n=11) | 4 | 3 | 2 | 2 | 0.95 |
| Present (n=22) | 8 | 4 | 4 | 6 |
| HER2 status | | | | | |
| Negative (n=23) | 10 | 7 | 3 | 3 | 0.021 |
| Positive (n=10) | 2 | 0 | 3 | 5 |
*Tumour any size with direct extension to the chest wall and/or to the skin
†Calculated using Fishers exact Test
Among the two cases of mucinous carcinoma, one of them was pre-menopausal, HR-positive, HER2-negative, Ki67-high, p53-negative tumour with intermediate grade histology, node-negativity and absent lympho-vascular invasion. The other case was post-menopausal, HR-negative, HER2-negative, Ki67-low, p53-negative tumour with histologic low grade features, lympho-vascular invasion and lymph node involvement.
Discussion
To optimise patient management in clinical practice, it is important to recognise patients as to who will or who will not benefit from particular therapies [15]. For this purpose, the established gene signatures being costly, the more widely available and cost-effective IHC biomarkers namely ER, PR and HER2 are used as surrogate approach [15]. The biological distinction between luminal A and B is provided by proliferation signature, including the genes CCNB1, MKI67 and MYBL2 of which MKI67 (encoding Ki67) is the most significant [43,44]. Cheang MCU et al., highlighted the clinical utility of the combined use of Ki67 with ER, PR and HER2 to distinguish Luminal-A from Luminal-B [28]. This separation is important to identify the high-risk for recurrence luminal-B patients who require additional chemotherapy from Luminal-A, for whom adjuvant endocrine therapy alone suffices [28]. Kobayashi T et al., and Lee SK et al., have shown that the combination of Ki67 and p53 in the IHC panel is more precise than Ki67 alone in predicting the prognosis for luminal breast disease [8,35]. The evaluation of breast cancer cases in the present study included both Ki67 and p53 IHC status along with the conventional biomarkers.
Studies [8-10,27-35] report the use of different Ki67 value cut-offs [Table/Fig-11], however 20% cut-off is confirmed as the best to stratify high-risk patients in luminal breast cancers [13,27,29,32,34,42]. It is reported that Ki67 positivity in >20%-50% of tumour cells confers high risk for recurrent disease [14]. At a 20% cut-off, 50% of all the cases in the present study as compared to 47% cases in the study by Shapochka DO et al., showed Ki67-high value [32]. Despite the variability in the Ki67 value cut-off’s, high Ki67 values are seen increasing in higher histologic grade of breast carcinoma [Table/Fig-12]. Studies have reported Ki67-high values in HER2-positive tumours [Table/Fig-13]. No significant correlation of Ki67-status with the tumour size was noted in the present study and study by Ding L et al., [33]. Though not statistically significant, a higher number (76%) of Ki67-high cases had positive nodes in the present study. Inic Z et al., observed lymph node positivity in 94% of Ki67-high cases [9].
Comparison of cut-off for Ki67-high value [8-10,27-35].
| Study | Ki67 cut-off value |
|---|
| Present study, | 20% |
| Acs B et al., [27] |
| Bustreo S et al., [29] |
| Ohara M et al., [34] |
| Shapochka DO et al., [32] |
| Soliman NA and Yussi SM, [10] | 15% |
| Plesan DMN et al., [30] |
| Inic Z et al., [9] | 14% |
| Shokouh TZ et al., [31] |
| Lee SK et al., [35] |
| Ding L et al., [33] |
| Cheang MCU et al., [28] | 13.25% |
| Kobayashi T et al., [8] | 10% |
Comparison: Relationship of Ki67 high value with histologic grade [10,30-33].
| Study | Total cases (n) | Ki67 high/positive and histologic grade |
|---|
| Ki67-high n (%) | Histologic grade n (%) | p-value |
|---|
| G1 | G2 | G3 |
|---|
| Present study | 50 | 25 (50) | 0 | 15 (60) | 10 (40) | 0.00006 |
| Plesan DMN et al., [30] | 100 | 45 (45) | 5 (11.11) | 40 (88.8) | n/a* |
| Shoukouh TZ et al., [31] | 566 | 225 (39.75) | 15 (6.7) | 127 (56.4) | 83 (36.9) | 0.001 |
| Shapochka DO et., al [32] | 62 | 29 (47) | r†=1 | <0.001 |
| Ding L et al., [33] | 257 | 193 (75) | 10 (5.18) | 96 (49.74) | 87 (45.08) | 0.001 |
| Soliman NA and Yussi SM, [10] | 107 | 36 (33.8) | 6 (17) | 14 (39) | 16 (44) | 0.00 |
*n/a: Not available; †r: correlation co-efficient
Comparison: Relationship of Ki67-high value with HER2 status positivity [30-33].
| Study | Total cases (n) | No. of HER2 +ve cases n (%) | No. of Ki67 high cases among HER2 +ve cases n (%) | p-value |
|---|
| Present study | 50 | 15 (30) | 12 (80) | 0.013 |
| Plesan DMN et al., [30] | 100 | 12 (12) | 11 (91.66) | n/a* |
| Shoukouh TZ et al., [31] | 566 | 111 (19.6) | 93 (83.8) | 0.001 |
| Ding L et al., [33] | 257 | 159 (61.8) | 137 (86.16) | <0.001 |
*n/a: Not available
Hanahan D and Weinberg RA have proposed eight distinctive and complementary hallmarks of cancer that enable tumour growth and differentiation [45]. Understanding the mechanism of the particular hallmarks helps in designing appropriate targeted therapies to treat the cancer. Dai X et al., identified the dominant hallmarks driving breast cancer heterogeneity, currently not used in molecular subtyping of breast cancer [4]. These include ‘resisting cell death’, ‘genome instability and mutation’ and ‘deregulating cellular energetics’; the use of these hallmark associated biomarkers namely BCL2, TP53, and VDR (vitamin-D receptor) respectively, will help to refine tumour classification specifically in terms of predictive value [4]. Resistance to medical treatments such as chemotherapy, hormone therapy and radiotherapy in 20-40% patients underscores the need for better knowledge of predictive factors of response to treatment [46-48]. It is found that adjuvant systemic therapy, especially with tamoxifen, along with radiotherapy is of less value for patients with TP53 mutated tumours [49]. Therapeutic strategies have focused on reactivation of wild-type function in the mutant p53 protein [50]. The anti-tumour function of small molecules that target p53 pathway are being examined in clinical trials [51,52].
Positive p53 status was more frequently observed in patients younger than 50 years and was significantly associated with pre-menopausal status in the present study. Similar results for age distribution [30] and menopausal status [53] was noted in the literature. As observed in 15 populations comprising both low- and high-risk (with respect to origin of the patients) for breast cancer, the frequency of somatic TP53 mutations ranged between 15-71% [36]. In the present study, the overexpression of p53 was encountered in 48% of cases studied, the result correlated with the reported data of 42% [30] and 44.96% [33]. Studies [30,33,35,54,55] report the occurrence of TP53 mutations more frequently in tumours with ductal and medullary histology, higher grade, large size, positive nodes and low hormone receptor status. This association is reported regardless of whether the p53 mutations were identified by IHC or other direct methods. Similar finding was noted in the present study [Table/Fig-14]. However, Song HS et al., reported significantly lower p53 positive status in lymph node metastasis cases [56]; both Yang P et al., and Song HS et al., reported no correlation of p53 overexpression with other clinico-pathological characteristics [6,56]. Overgaard J et al., demonstrated that nodal status and TP53 mutation expressed independent poor prognostic significance for Overall-Survival (OS) and Disease-Free Survival (DFS) [55]; and menopausal status had independent significance for OS [55]. Pharoah PD et al., in a meta-analysis of 16 studies and Olivier M et al., in the investigation of 1,794 breast cancer patients confirmed that TP53 mutation is an independent negative prognostic factor conferring poorer OS and DFS in breast cancer [19,54]. Ding L et al., noted significantly higher p53 overexpression in HER2-positive patients than in HER2-negative patients; the reverse result was observed in the present study [33].
Comparison: Percentage of p53 positive cases with ductal/medullary histology, high grade, large size, positive nodes and low hormone receptor (HR) status.
| Total cases (n) | p53 mutation positive (%) | Histology [Ductal or medullary carcinoma] (%) | Histologic grade [Intermediate- High] (%) | Large size [>2 cm] (%) | Positive nodes (%) | HR status low (%) |
|---|
| Present study | 50 | 48 | 100 | 96 | 87.5 | 83.3 | 67 |
| Olivier M et al., [54] | 1794 | 17 | 90.43 | 99 | 80.2 | 48.6 | 52 |
| Plesan DMN et al., [30] | 100 | 42 | 95.24 | 71 (G3) | 66.64 | n/a | 61.9 |
| Ding L et al., [33] | 258 | 44.96 | 85.34 | 95.69 | n/a* | n/a* | n/a* |
| Overgaard J et al., [55] | 294 | 23 | 94.2 | 91.3 | 65.2 | 59.4 | 46.5 |
| Lee SK et al., [35] | 7739 | 28.77 | n/a* | s/a† | s/a† | n/a* | s/a† |
*n/a: Not available; †s/a: Statistically associated
Literature review of IHC based molecular-subtyping (including Ki67 in the panel) of breast carcinoma reveals that there is no uniformity in the sub-type to which the HER2-positive HR-positive cases are assigned; they are categorised as either HER2-like [5] or luminal-B/luminal-B hybrid [28,31,32]. Statistically significant association of molecular subtypes with Ki67-positivity is reported in the present study and other studies [10]. Whole-genome analysis has identified the frequency of TP53 mutation to be higher in Luminal-B (29%) than in Luminal-A (12%) breast cancers [57]; also Luminal-A tumours are reported to be less proliferative and have lower rate of p53 overexpression [8,44] suggesting the use of p53-status to distinguish Luminal-A from Luminal-B breast cancers. In the present study, 41% of Luminal-A and 50% of Luminal-B patients showed p53 overexpression which was not statistically significant. However, the Luminal-A tumours were significantly less proliferative. The results of this study showed that Luminal-A patients tend to be older, postmenopausal and had node negative tumours; similar to that reported in other studies [26]. Unlike other study data [26] there was no significant association of lympho-vascular invasion with Luminal-B subtype in our study. It is reported that Luminal-A subtype presents a significantly lower risk of early tumour recurrence [14]. Luminal-A tumours are significantly associated with grade 1 and 2 histology in the present study. Same finding is reported in the literature [10].
Literature data [27,32] reveals that HR-positive tumours are the most common, followed by Basal-like and HER2-positive HR-negative type [Table/Fig-15]. Studies [8,34,35] have evaluated the status of both Ki67 and p53 in luminal and/or HR-positive breast cancers. Ohara M et al., assessed 308 luminal-type breast cancer patients and confirmed the prognostic utility of Ki67 status, whereas no prognostic significance for p53 was revealed [34]. In their study of luminal-type breast cancers, Kobayashi T et al., observed that combined Ki67-p53 status was more accurate than Ki67 alone in predicting patient outcome [8]. Similar to this study, in the present study the HR-positive tumours with ‘favourable’ Ki67-low and p53-negative phenotype showed lower frequencies of higher nuclear grades and HER2 positivity than the ‘unfavourable’ phenotype tumours. Kobayashi T et al., noted all HER2-positive tumours to be unfavourable phenotype tumours, whereas in the present study 80% of HER2-positive were of the unfavourable phenotype tumours [8]. In a somewhat similar manner to Kobayashi T et al., Lee SK et al., in their study of 7,739 cases of invasive breast carcinoma, classified only luminal (HR positive, HER2 negative) cases into ‘low-risk’ Ki67-low, p53 negative subtype and ‘high-risk’ Ki67-low p53-positive, Ki67-high p53-negative and Ki67-high p53-positive subtypes [8,35]. They determined 10% IHC nuclear staining as the suitable p53 overexpression cut-off value to predict OS and DFS especially in Luminal ER and PR positive breast cancer. They concluded that the combined Ki67-p53 status was superior to that of either p53 or Ki67 alone in the prediction of DFS.
Comparison of the common subtypes of breast carcinoma [27,32].
| Study | No. of cases | HR-Positive n (%) | HER2+ve HR-ve n (%) | Basal-like n (%) |
|---|
| Present study | 50 | 33 (66) | 5 (10) | 12 (24) |
| Acs B et al., [27] | 120 | 80 (66.67) | 14 (11.67) | 26 (21.67) |
| Shapochka DO et al., [32] | 62 | 43 (69) | 7 (11) | 12 (20) |
HR: Hormone receptor
Limitation(s)
The limited number of cases evaluated and lack of follow-up period in the present study precludes the confirmation of observations and identification of any other associations that may have existed. Yet, available methodology and cut-off values for Ki67 and p53 IHC analyses lack standardisation.
Conclusion(s)
The present study identified a statistical significance for the association of Ki67 status with the histologic grade, HER2 status and clinically defined-therapy oriented molecular subtypes. In the HR-positive cases, the evaluation of combined Ki67-p53 status has provided significant correlation with menopausal status, histologic grade, lymph node status and HER2 status. Considering the results of this study and the literature data revealing significant number (66 to 70%) of breast cancers expressing a HR, the use of combined Ki67-p53 status will play a significant role in discriminating the HR-positive patients who could benefit from aggressive treatment, thus optimising the cost-benefit ratio. Prospective studies with follow-up in a larger population will be useful to assess the impact of stratifying luminal cases into favourable and unfavourable types and thereby enlighten the role of p53 in therapeutic decisions. Further, inconsistencies in the methodology and cut-off for reporting Ki67 and p53 status underscores the need for a uniform training for all the researchers so that substantial and meaningful data can be pooled to generate cost-effective treatment decisions especially in HR positive breast cancers.
*Tumour any size with direct extension to the chest wall and/or to the skin
HR: Hormone receptor; +: Positive; -: Negative
*Tumour any size with direct extension to the chest wall and/or to the skin
†Calculated using Fishers exact Test
*Tumour any size with direct extension to the chest wall and/or to the skin
†Calculated using Fishers exact Test; **Calculated using Chi square test with Yates’ correlation
*Calculated using Chi square test with Yates’ correlation
†Calculated using Fishers exact Test
HR: Hormone receptor; +: Positive; -: Negative
*Calculated using Chi square test with Yates’ correlation
†Calculated using Fishers exact Test
HR: Hormone receptor; +: Positive; -: Negative
*Tumour any size with direct extension to the chest wall and/or to the skin
†Calculated using Fishers exact Test
*n/a: Not available; †r: correlation co-efficient
*n/a: Not available
*n/a: Not available; †s/a: Statistically associated
HR: Hormone receptor
[1]. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countriesCA Cancer J Clin 2018 68(6):394-424.10.3322/caac.2149230207593 [Google Scholar] [CrossRef] [PubMed]
[2]. Trends over time for all sites and on selected sites of cancer & projection of burden of cancer. In: Three-year report of population based cancer registries 2012-2014. Bengaluru: NCDIR-NCRP(ICMR); 2016. p. 89-125 [Google Scholar]
[3]. Andre F, Pusztai L, Molecular classification of breast cancer: Implications for selection of adjuvant chemotherapyNat Clin Pract Oncol 2006 3(11):621-32.10.1038/ncponc063617080180 [Google Scholar] [CrossRef] [PubMed]
[4]. Dai X, Xiang L, Li T, Bai Z, Cancer hallmarks, biomarkers and breast cancer molecular subtypesJ Cancer 2016 7(10):1281-94.10.7150/jca.1314127390604 [Google Scholar] [CrossRef] [PubMed]
[5]. Hortobagyi GN, Connolly JL, D’Orsi CJ, Edge SB, Mittendorf EA, Rugo HS, Breast. In: Amin MB, editorsAJCC Cancer Staging Manual 2017 8th edSwitzerlandSpringer:589-628.10.1007/978-3-319-40618-3_48 [Google Scholar] [CrossRef]
[6]. Yang P, Du CW, Kwan M, Liang SX, Zhang GJ, The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasisSci Rep 2013 3(2246):01-06.10.1038/srep0224623873310 [Google Scholar] [CrossRef] [PubMed]
[7]. Dowsett M, Nielsen TO, A’Hem R, Bartlett J, Coombes RC, Cuzick J, Assessment of Ki67 in breast cancer: Recommendations from the international Ki67 in breast cancer working groupJ Natl Cancer Inst 2011 103(22):01-09.10.1093/jnci/djr39321960707 [Google Scholar] [CrossRef] [PubMed]
[8]. Kobayashi T, Iwaya K, Moriya T, Yamasaki T, Tsuda H, Yamamoto J, A simple immunohistochemical panel comprising 2 conventional markers, Ki67 and p53, is a powerful tool for predicting patient outcome in luminal-type breast cancerBMC Clin Pathol 2013 13(5):01-11.10.1186/1472-6890-13-523384409 [Google Scholar] [CrossRef] [PubMed]
[9]. Inic Z, Zegarac M, Inic M, Markovic I, Kozomara Z, Djurisic I, Difference between luminal A and luminal B subtypes according to Ki-67, tumor size, and progesterone receptor negativity providing prognostic informationClin Med Insights Oncol 2014 8:107-11.10.4137/CMO.S1800625249766 [Google Scholar] [CrossRef] [PubMed]
[10]. Soliman NA, Yussi SM, Ki-67 as a prognostic marker according to breast cancer molecular subtypeCancer Biol Med 2016 13(4):496-504.10.20892/j.issn.2095-3941.2016.006628154782 [Google Scholar] [CrossRef] [PubMed]
[11]. Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA, Ki67 in breast cancer: Prognostic and predictive potentialLancet Oncol 2010 11(2):174-83.10.1016/S1470-2045(09)70262-1 [Google Scholar] [CrossRef]
[12]. Denkert C, Budczies J, von Minckwitz G, Wienert S, Loibl S, Klauschen F, Strategies for developing Ki67 as a useful biomarker in breast cancerThe Breast 2015 24:S67-72.10.1016/j.breast.2015.07.01726283598 [Google Scholar] [CrossRef] [PubMed]
[13]. Leung SCY, Nielsen TO, Zabaglo L, Arun I, Badve SS, Bane AL, Analytical validation of a standardized scoring protocol for Ki67: Phase 3 of an international multi centre collaborationNPJ Breast Cancer 2016 2(16014):01-09.10.1038/npjbcancer.2016.1428721378 [Google Scholar] [CrossRef] [PubMed]
[14]. Joensuu K, Leidenius M, Kero M, Andersson LC, Horwitz KB, Heikkila P, ER, PR, HER2, Ki67 and CK5 in early and late relapsing breast cancer- Reduced CK5 expression in metastasesBreast Cancer (Auckl) 2013 7:23-34.10.4137/BCBCR.S1070123514931 [Google Scholar] [CrossRef] [PubMed]
[15]. Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Tailoring therapies- improving the management of early breast cancer: St Gallen international expert consensus on the primary therapy of early breast cancer 2015Ann Oncol 2015 26(8):1533-46.10.1093/annonc/mdv22125939896 [Google Scholar] [CrossRef] [PubMed]
[16]. Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ, Thresholds for therapies: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2009Ann Oncol 2009 20(8):1319-29.10.1093/annonc/mdp32219535820 [Google Scholar] [CrossRef] [PubMed]
[17]. Levine AJ, Momand J, Finlay CA, The p53 tumour suppressor geneNature 1991 351(6326):453-56.10.1038/351453a02046748 [Google Scholar] [CrossRef] [PubMed]
[18]. Varna M, Bousquet G, Plassa LF, Bertheau P, Janin A, TP53 status and response to treatment in breast cancersJ Biomed Biotechnol 2011 2011:01-09.10.1155/2011/28458421760703 [Google Scholar] [CrossRef] [PubMed]
[19]. Pharoah PD, Day NE, Caldas C, Somatic mutations in the p53 gene and prognosis in breast cancer: A meta-analysisBr J Cancer 1999 80(12):1968-73.10.1038/sj.bjc.669062810471047 [Google Scholar] [CrossRef] [PubMed]
[20]. Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics dataHum Mutat 2016 37(9):865-76.10.1002/humu.2303527328919 [Google Scholar] [CrossRef] [PubMed]
[21]. Reich NC, Levine AJ, Growth regulation of a cellular tumour antigen, p53, in nontransformed cellsNature 1984 308(5955):199-201.10.1038/308199a06366574 [Google Scholar] [CrossRef] [PubMed]
[22]. Bartek J, Bartkova J, Lukas J, Statskova Z, Vojtesek B, Lane DP, Immunohistochemical analysis of the p53 oncoprotein on paraffin sections using a series of novel monoclonal antibodiesJ Pathol 1993 169(1):27-34.10.1002/path.17116901068433213 [Google Scholar] [CrossRef] [PubMed]
[23]. Oren M, Regulation of the p53 tumour suppressor proteinJ Biol Chem 1999 274(51):36031-34.10.1074/jbc.274.51.3603110593882 [Google Scholar] [CrossRef] [PubMed]
[24]. Norberg T, Lennerstrand J, Inganas M, Bergh J, Comparison between p53 protein measurements using the luminometric immunoassay and immunohistochemistry with detection of p53 gene mutations using cDNA sequencing in human breast tumoursInt J Cancer 1998 79(4):376-83.10.1002/(SICI)1097-0215(19980821)79:4<376::AID-IJC12>3.0.CO;2-3 [Google Scholar] [CrossRef]
[25]. Fitzgibbons PL, Page DL, Weaver D, Thor AD, Allred DC, Clark GM, Prognostic factors in breast cancer. College of American pathologists consensus statement 1999Arch Pathol Lab Med 2000 124(7):966-78. [Google Scholar]
[26]. Inwald EC, Koller M, Schalke KM, Zeman F, Hofstadter F, Gerstenhauer M, 4-IHC classification of breast cancer subtypes in a large cohort of a clinical cancer registry: Use in clinical routine for therapeutic decisions and its effect on survivalBreast Cancer Res Treat 2015 153:647-58.10.1007/s10549-015-3572-326369534 [Google Scholar] [CrossRef] [PubMed]
[27]. Acs B, Zambo V, Vizkeleti L, Szasz AM, Madaras L, Szentmartoni G, Ki-67 as a controversial predictive and prognostic marker in breast cancer patients treated with neoadjuvant chemotherapyDiagn Pathol 2017 12(20):01-12.10.1186/s13000-017-0608-528222768 [Google Scholar] [CrossRef] [PubMed]
[28]. Cheang MCU, Chia SK, Voduc D, Gao D, Leung S, Snider J, Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancerJ Natl Cancer Inst 2009 101(10):736-50.10.1093/jnci/djp08219436038 [Google Scholar] [CrossRef] [PubMed]
[29]. Bustreo S, Abate SO, Cassoni P, Donadio M, Airoldi M, Pedani M, Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: A large case series study with a long term follow-upBreast Cancer Res Treat 2016 157(2):363-71.10.1007/s10549-016-3817-927155668 [Google Scholar] [CrossRef] [PubMed]
[30]. Plesan DMN, Georgescu CV, Patrana N, Plesan C, Stoica D, Immunohistochemical study of p53 and Ki67 in a group of patients with mammary carcinomaRom J Morphol Embryo 2010 51(3):459-65. [Google Scholar]
[31]. Shokouh TZ, Ezatollah A, Barand P, Interrelationships between Ki67, Her2/neu, p53, ER, and PR status and their associations with tumour grade and lymph node involvement in breast carcinoma subtypes: Retrospective-observational analytical studyMedicine 2015 94(32):01-06.10.1097/MD.000000000000135926266392 [Google Scholar] [CrossRef] [PubMed]
[32]. Shapochka DO, Zaletok SP, Gnidyuk M I, Relationship between NF-KB, ER, PR, HER2/Neu, Ki67, P53 expression in human breast cancerExp Oncol 2012 34(4):358-63. [Google Scholar]
[33]. Ding L, Zhang Z, Xu Y, Zhang Y, Comparative study of Her-2, p53, Ki-67 expression and clinicopathological characteristics of breast cancer in a cohort of northern China female patientsBioengineered 2017 8(4):383-92.10.1080/21655979.2016.123510128075663 [Google Scholar] [CrossRef] [PubMed]
[34]. Ohara M, Matsuura K, Akimoto E, Noma M, Doi M, Nishizaka T, Prognostic value of Ki67 and p53 in patients with estrogen receptor-positive and human epidermal growth factor receptor 2-negative breast cancer: Validation of the cut-off value of the Ki67 labelling index as a predictive factorMol Clin Oncol 2016 4(4):648-54.10.3892/mco.2016.77627073684 [Google Scholar] [CrossRef] [PubMed]
[35]. Lee SK, Bae SY, Lee JH, Lee HC, Yi H, Kil WH, Distinguishing low-risk luminal A breast cancer subtypes with Ki-67 and p53 is more predictive of long-term survivalPLoS One 2015 10(2-3):01-14.10.1371/journal.pone.012465826241661 [Google Scholar] [CrossRef] [PubMed]
[36]. Hill KA, Sommer SS, p53 as a mutagen test in breast cancerEnviron Mol Mutagen 2002 39:216-27.10.1002/em.1006511921192 [Google Scholar] [CrossRef] [PubMed]
[37]. Lakhani SR, Ellis IO, Schnitt SJ, Tan PY, Vijver MJ, WHO Classification of Tumours of the Breast 2012 4th edLyon, FranceIARC Press:1-240. [Google Scholar]
[38]. Bloom HJ, Richardson WW, Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 yearsBr J Cancer 1957 11(3):359-77.10.1038/bjc.1957.4313499785 [Google Scholar] [CrossRef] [PubMed]
[39]. Elston CW, Ellis IO, Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow upHistopathology 1991 19(5):403-10.10.1111/j.1365-2559.1991.tb00229.x1757079 [Google Scholar] [CrossRef] [PubMed]
[40]. Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badave S, American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancerArch Pathol Lab Med 2010 134(7):E48-72. [Google Scholar]
[41]. Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of American pathologists clinical practice guideline focused updateArch Pathol Lab Med 2018 142:1364-82.10.5858/arpa.2018-0902-SA29846104 [Google Scholar] [CrossRef] [PubMed]
[42]. Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013Ann Oncol 2013 24(9):2206-23.10.1093/annonc/mdt30323917950 [Google Scholar] [CrossRef] [PubMed]
[43]. Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Distinctive gene expression patterns in human mammary epithelial cells and breast cancersProc Natl Acad Sci USA 1999 96(16):9212-17.10.1073/pnas.96.16.921210430922 [Google Scholar] [CrossRef] [PubMed]
[44]. Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, The molecular portraits of breast tumours are conserved across microarray platformsBMC Genomics 2006 7:9610.1186/1471-2164-7-9616643655 [Google Scholar] [CrossRef] [PubMed]
[45]. Hanahan D, Weinberg RA, Hallmarks of cancer: The next generationCell 2011 144(5):646-74.10.1016/j.cell.2011.02.01321376230 [Google Scholar] [CrossRef] [PubMed]
[46]. Robertson JRR, Cussac AL, Rolski J, Feltl D, Dewar J, Macpherson E, Activity of fulvestrant 500 mg versus anastrozole 1 mg as first-line treatment for advanced breast cancer: Results from the FIRST studyJ Clin Oncol 2009 27(27):4530-35.10.1200/JCO.2008.21.113619704066 [Google Scholar] [CrossRef] [PubMed]
[47]. Miles DW, Chan A, Dirix LY, Cortés J, Pivot X, Tomczak P, Phase III study of bevacizumab plus docetaxel compared with placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancerJ Clin Oncol 2010 28(20):3239-47.10.1200/JCO.2008.21.645720498403 [Google Scholar] [CrossRef] [PubMed]
[48]. Andersson M, Lidbrink E, Bjerre K, Wist E, Enevoldsen K, Jensen AB, Phase III randomised study comparing docetaxel plus trastuzumab with vinorelbine plus trastuzumab as first-line therapy of metastatic or locally advanced human epidermal growth factor receptor-2 positive breast cancer: The HERNATA studyJ Clin Oncol 2011 29(3):264-71.10.1200/JCO.2010.30.821321149659 [Google Scholar] [CrossRef] [PubMed]
[49]. Bergh J, Norberg T, Sjogren S, Lindgren A, Holmberg L, Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapyNat Med 1995 1(10):1029-34.10.1038/nm1095-10297489358 [Google Scholar] [CrossRef] [PubMed]
[50]. Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomasNature 2007 445(7128):656-60.10.1038/nature0552917251933 [Google Scholar] [CrossRef] [PubMed]
[51]. Lehmann S, Bykov VJN, Ali D, Andrén O, Cherif H, Tidefelt U, Targeting p53 in vivo: A first in human study with p53 targeting compound APR-246 in refractory hematologic malignancies and prostate cancerJ Clin Oncol 2012 30(29):3633-39.10.1200/JCO.2011.40.778322965953 [Google Scholar] [CrossRef] [PubMed]
[52]. Zawacka PJ, Selivanova G, Pharmacological reactivation of p53 as a strategy to treat cancerJ Intern Med 2015 277(2):248-59.10.1111/joim.1233625495071 [Google Scholar] [CrossRef] [PubMed]
[53]. Al-Moundhri M, Nirmala V, Al-Mawaly K, Ganguly S, Burney I, Rizvi A, Significance of p53, Bcl-2, and Her-2/neu protein expression in Omani Arab females with breast cancerPathol Oncol Res 2003 9(4):226-31.10.1007/BF0289338214688828 [Google Scholar] [CrossRef] [PubMed]
[54]. Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancerClin Cancer Res 2006 12(4):1157-67.10.1158/1078-0432.CCR-05-102916489069 [Google Scholar] [CrossRef] [PubMed]
[55]. Overgaard J, Yilmaz M, Guldberg P, Hansen LL, Alsner J, TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancerActa Oncol 2000 39(3):327-33.10.1080/02841860075001309610987229 [Google Scholar] [CrossRef] [PubMed]
[56]. Song HS, Do YR, Kang SH, Jeong KY, Kim YS, Prognostic significance of immunohistochemical expression of p53 gene product in operable breast cancerCancer Res Treat 2006 38(4):218-23.10.4143/crt.2006.38.4.21819771246 [Google Scholar] [CrossRef] [PubMed]
[57]. Perou CM, Comprehensive molecular portraits of human breast tumours. The cancer genome atlas networkNature 2012 490(7418):61-70.10.1038/nature1141223000897 [Google Scholar] [CrossRef] [PubMed]