Cervical cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer death in women worldwide [1]. Despite the implementation of Pap test that has successfully brought dramatic reduction in the incidence and mortality worldwide caused by cervical cancer, the false positive and false negative rates of this test are significant [2], proving diagnostic limitation of the Pap test and also possibility of under or over-treatment. Also, in histopathology of cervical biopsies too, there could be intra-observer and inter-observer diagnostic discrepancies even among panel of pathologists reviewing the same slides [3]. Hence, various Immunohistochemistry (IHC) biomarkers are being evaluated to differentiate pre-cancerous lesions and uterine cervical carcinoma in histopathology. p16 IHC is an established method to differentiate the grades of CIN and in this study, we strengthen this fact by using a combined scoring method [4] and its association with Human Papilloma Virus (HPV) DNA. A comment on the prevalent serotypes of HPV in India and possible alteration in HPV vaccination in Indian population is also added. The aim was to study p16 IHC in spectrum of CIN and to access the possible utility of p16 IHC in differentiating CIN1/L-SIL from CIN2,3/H-SIL and to measure strength of association of p16 IHC with HPV DNA status in various grades of cervical intra-epithelial lesions, wherever available.
Materials and Methods
Study design was retrospective and hospital based and was conducted from January 2016 to March 2017 for a period of 15 months in Pune, India. The study was actually an IHC study done in Dept. of pathology, BJGMC and the HPV DNA results of same cases from Dept. of microbiology, BJGMC were used. Ethical Committee Clearance (No. D-1214130-130) was taken for the study. The study included 50 consecutive biopsies having a diagnosis of cervical intraepithelial neoplasia, from January 2015 to December 2016. The power of the study was 0.8. The below mentioned inclusion and exclusion criteria were applied. All cervical biopsies with light microscopy diagnosis of CIN were included. All cervical biopsies with light microscopy diagnosis other than CIN were excluded out of the study which included non-dysplastic cases and malignant squamous cell carcinoma cases.
The demographic data was collected followed by cervical biopsies of the patients for histopathological examination. For microscopy, detailed sectioning of the gross specimen was done and multiple sections from suspected areas were taken and embedded in paraffin blocks. Routine haematoxylin and eosin staining was done followed by IHC.
Immunohistochemistry (IHC) Procedure
The immunohistochemical staining with p16 was done using the two step polymer method of IHC. The standard operating procedure of IHC staining (SOP) of the institute was followed. Positive control for IHC was p16 positive squamous cell carcinoma of cervix. The staining positivity, intensity, pattern (focal or diffuse) of IHC were observed and correlated with the histopathological examination of the same cases. A Scoring was given to each slide stained with IHC for p16 according to Vinyuvat S et al., [4].
The scoring was given according to the following criteria:
Percentage of positive cells: 0: <5% positive cells, 1: 5-49% positive cells.2: 50-80% positive cells. 3: >80% positive cells.
Intensity of reaction: 0: No reaction, 1: weak reaction, 2: variable (both weak and strong) reaction, 3: Strong reaction
Cellular reaction pattern: 0: No reaction, 1: Focal reaction, 2: Diffuse reaction
A diffuse reaction was defined as a positive reaction greater than 1 low power field (100x magnification) [4]. A total score was then given on the basis of all three parameters with a range of 0 to 8. All cases with score of 4 and greater were considered positive for p16. The data obtained were analysed statistically, to differentiate between different grades of cervical intraepithelial neoplasia (CIN 1, 2 and 3) by the use of histopathology and p16 IHC investigation.
Due to a resource limited setting, the HPV status of the patients wherever available were included in the study, which accounted for 41 out of 50 cases. HPV detection in our institute is done by the Abbott m2000rt High Risk HPV (HR-HPV) Assay [5]. The results were reported as Detected (HPV 16/ HPV 18/ Other HR HPV) or not detected. Other HR-HPV subtypes included HPV 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68.
Statistical Analysis
This is an observational study and hence the results were represented in percentage forms. Graphical representation was done wherever possible. The SPSS 18.0 software was used for statistical analysis and Fisher’s-exact test was used for comparisons between the groups for p16 IHC and CIN subtypes. The strength of association between p16 IHC and HPV DNA was calculated using Phi coefficient test.
Results
Mean age (yr) in biopsies with CIN (Overall) was 35.9 years, in biopsies with CIN1: 35.9 years, in CIN2 was 37.1 years and in CIN3 was 32.3 years. The Age Range of the case was 24-50 years.
p16 IHC in CIN
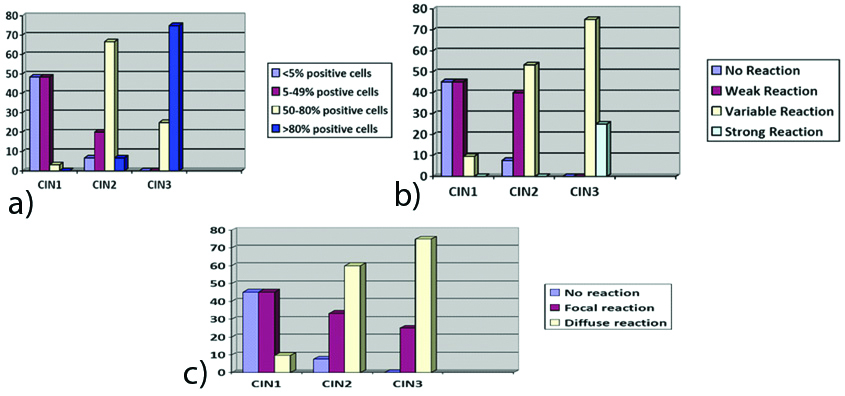
p16 was scored according to the combined scoring system as proposed by Vinyuvat S et al., according to the following criteria: Percentage of positive cells, Intensity of reaction and Cellular reaction pattern [4]. [Table/Fig-1,2,3 and 4] show the IHC results of percentage of p16 positive cells, intensity of p16 reaction and cellular reaction pattern. It was found that all the above parameters i.e., percentage of positive cells, intensity of p16 IHC reaction as well as the cellular reaction pattern increased as the grade of dysplasia increased.
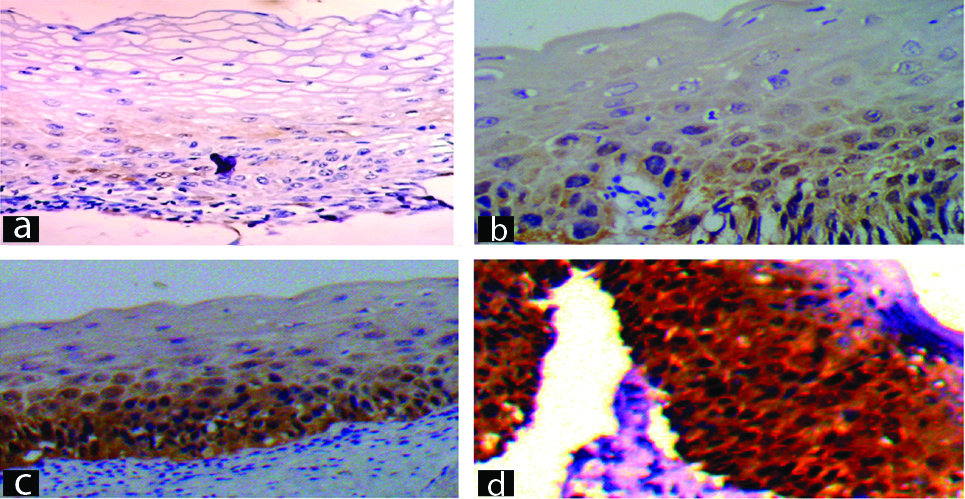
Persentage of positive cells on p16 IHC.
a) Score 0: <5% cells positive on p16 IHC (p16 IHC, 400X); b) Score : 5-50% cells positive on p16 IHC (p16 IHC, 400X); c) Score 2: 50-80% cells positive on p16 IHC (p16 IHC, 400X); d) Score 3: >80% cells positive on p16 IHC (p16 IHC, 400X).
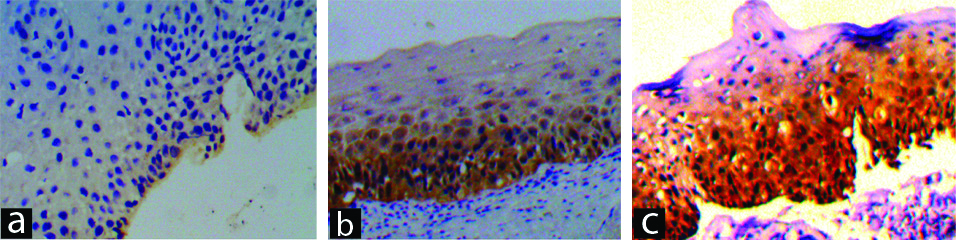
Intensity of reaction of p16 IHC staining.
a) Score 1: Weak intensity of reaction on p16 IHC (p16 IHC, 100X). b) Score 2: Variable (both weak and strong) intensity of reaction on p16 IHC (p16 IHC, 100X). c) Score 3: Strong intensity of reaction on p16 IHC (p16 IHC, 100X).
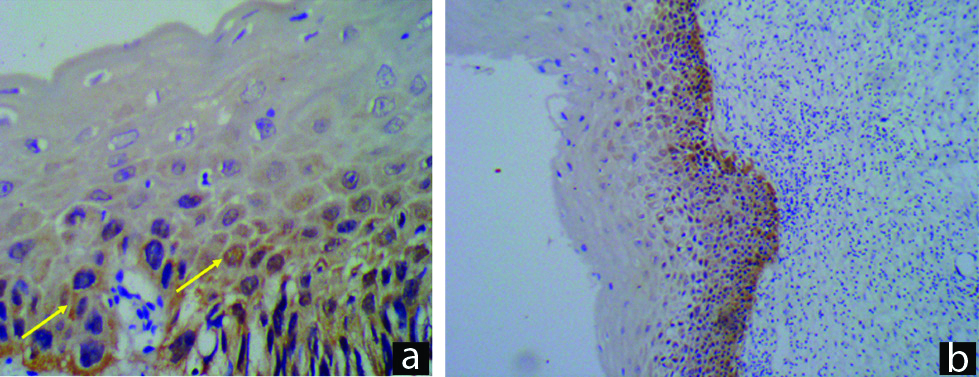
Cellular reaction pattern.
a) Score 1: Focal reaction pattern, p16, IHC (p16 IHC, 400X), b) Score 2: Diffuse reaction pattern on p16 IHC (p16 IHC, 100X).
a) IHC results for percentage of p16IHC, percentage of positive cells (X axis: grades of CIN, Y axis percentage of cases); b) IHC results for percentage of p16 IHC, intensity of p16 reaction (X axis: grades of CIN, Y axis percentage of cases); c) IHC results for percentage of p16 IHC, cellular reaction pattern to p16 IHC in different grades of CIN (X axis: grades of CIN, Y axis percentage of cases).
Correlation of Histopathology (HPE) Results with p16 IHC Results
As seen in [Table/Fig-1,2,3 and 4], 24% of CIN1 cases, 80% of CIN2 cases and 100% of CIN3 cases were positive for p16 IHC. Thus, it can be concluded that the positivity to p16 IHC increased from CIN1 to CIN3 i.e., it increased with increasing grades of dysplasia. Of the total 50 cases of cervical intraepithelial neoplasia, 44% were positive for p16 IHC.
IHC in Differentiating Various Grades of CIN
The present study as seen in [Table/Fig-5] showed that while differentiating between H-SIL versus (vs) L-SIL and CIN2 vs CIN1, p-value was <0.05 i.e., the result was significant. Therefore, it can be concluded that p16 was helpful in differentiating H-SIL vs L-SIL and CIN2 vs CIN1 however while differentiating between CIN3 vs CIN2, the p-value was >0.05 i.e., the result was not significant. Therefore, it can be concluded that p16 was not helpful in differentiating CIN3 and CIN2.
Sensitivity, specificity, positive predictive value and negative predictive value of p16.
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|
| CIN 2,3 vs CIN 1(H-SIL vs L-SIL) | 84.2 | 80.6 | 72.7 | 89.3 | 82 |
| CIN 2 vs CIN 1 | 80 | 80.6 | 66.7 | 89.3 | 80.4 |
| CIN 3 vs CIN 2 | 100 | 20 | 25 | 100 | 36.8 |
Correlation of Intracellular p16 pattern and Histopathological Diagnosis (HPE)
The study also correlated intracellular p16 IHC pattern with histopathological diagnosis. p16 IHC is considered as positive when the cytoplasm and/or nuclei of cells stain positive. Nuclear positivity, whenever present did not vary but cytoplasmic positivity varied from weak to strong positivity and increased from weak to strong with increasing grades of dysplasia.
Correlation of Histopathological Diagnosis, p16 IHC Positivity and HPV Positivity
p16 is considered as an indirect marker for HPV. Out of 25 cases of CIN1, 16% (n=4) showed p16 IHC positivity but 76% (n=19) showed High risk HPV (HR-HPV) positivity. Out of 12 cases of CIN2, 83.4% (n=10) showed p16 IHC positivity and 91.7% (n=11) showed HR-HPV positivity. Out of 4 cases of CIN3, all cases showed p16 IHC positivity and HR-HPV positivity. Only those cases that had HPV testing done are included and therefore value of ‘n’ is different. Also, a single case can be positive for more than one HR-HPV subtype. In the present study, the phi coefficient of association between p16 IHC and HPV DNA for L-SIL cases (CIN1) was -0.01 and the phi coefficient of association between p16 IHC and HPV DNA for H-SIL cases (CIN1) was 0.74. It can therefore be concluded that p16 IHC and High risk HPV status showed strong association in H-SIL (CIN2 and CIN3) cases but no or negligible association in L-SIL (CIN1) cases.
Correlation of CIN and High Risk HPV Subtypes
Considering the total cases of CIN, Other HR-HPV were the most common with 63.4% cases showing positivity, which was followed by HPV 16 (36.6% positivity) and HPV 18 (4.9% positivity). A 17.1% cases (n=7) were negative for all types of High risk HPV.
Discussion
p16 or p16/INK4a is a Cyclin Dependant Kinase (CDK) inhibitor 2(CDKN2) that antagonises the functions of CDK 4 and CDK 6, which are activated by binding with cyclin D1. Interestingly, p16 is upregulated in HPV-related lesions and this increase in p16 is actually a compensatory mechanism to inhibit the growth of CDK4 and CyclinD that are increased due to HPV replication. Therefore, p16 can be considered as an indirect marker for presence of altered HR-HPV and growth cycle transformation [6].
Age Distribution of Cases
The mean ages among the various grades of CIN were CIN1=35.9 years, CIN2=37.1 years and CIN3=32.3 years. There is a conventional hypothesis that CIN progresses from lower grades to higher grades i.e., from CIN1 to CIN3. However, the lower mean age in CIN3 cases, proves otherwise. This lower mean age maybe due to unknown confounding factor or it may be due to HPV16 infection in these cases. There is special mention in literature by Sideri M et al., [7], that HPV 16 infection causes direct progression to CIN3, causing a lower mean age in such cases and other High risk HR-HPV follow the conventional rule. In the present study, 3 out of 4 cases of CIN3, were positive for HPV 16 infection, thus explaining the lower mean age. In the present study, 1 case was positive for other HR- HPV, but was of the age of 30 years, which does not correlate with the study done by Sideri M et al., [7].
p16 IHC in CIN
Comparison of p16 IHC positivity in the CIN spectrum of various studies which used similar or different parameters showed an increasing positivity with increasing grade of CIN. [Table/Fig-6,7] However, the wide distribution of such studies in various grades of CIN can be attributed to different type of antibody used in IHC, different standard operating procedures for IHC staining or unavailability of a standard scoring system in cases of p16 in CIN cases [19,20]. A scoring system similar to the one used in the present study, will definitely help to standardise results of p16 IHC.
Comparison of p16 IHC positivity in spectrum of CIN of various studies using similar parameters with present study [8-10].
| p16 positivity |
|---|
| CIN 1 | CIN 2 | CIN 3 |
|---|
| Queiroz C et al., [8] (2006) | 53.3% | 70% | 93.4% |
| Galgano MT et al., [9] (2010) | 58.1% | 85.3% | 99.2% |
| Alshenawy HA et al., [10] (2014) | 27% | 54% | 85% |
| Present study (2019) | 24% | 80% | 100% |
Comparison of p16 IHC positivity in spectrum of CIN of various studies using different parameters with present study [11-18].
| Parameter of p16 IHC used | p16 positivity |
|---|
| CIN 1/ LSIL | CIN 2 | CIN 3 |
|---|
| Keating JT et al., [11] (2001) | Strong positive | 37.5% | 70.2% (H-SIL) |
| Klaes R et al., [12] (2001) | Diffuse positivity | 61% | 100% | 100% |
| Agoff SN et al., [13] (2003) | p16 >75% positive | 12% | 35% | 73% |
| Wang SS et al., [14] (2004) | Diffuse positivity | 36% | 63% | 100% |
| Murphy N et al., [15] (2005) | p16 >50% positive | 60% | 52% | 59% |
| Nam EJ et al., [16] (2008) | Nuclear positivity | 27% | 37% | 62% |
| Liu HQ et al., [17] (2015) | Percentage of positive cells | 30% | 56% | 82% |
| Zhang G et al., [18] (2015) | Pattern of staining | 50% | 100% | 100% |
| Present study (2019) | Vinyuvat S et al., [4] scoring | 24% | 80% | 100% |
Diffuse p16 positive CIN1 cases: There were a significant number of such cases, in all of the above mentioned studies, this subset of CIN1 cases are at higher risk of progression to H-SIL/CIN2-3. These cases also need a closer follow-up.
Intracellular p16 Pattern and HPE
In the present study, it was found that low grade SIL showed weak cytoplasmic p16 IHC positivity while strong cytoplasmic p16 IHC positivity was seen in higher grades of CIN and this feature can be used as an adjunct in differentiating between low and high grades of CIN. This feature was also seen in studies done by Quieroz C et al., and Lesnikova I et al., [8,21]. The cellular positivity of p16 may be due to a post- transcriptional modification or simply overproduction of p16 forcing its transfer to the cytoplasm as proposed by Murphy N et al., [22].
Comparison of Spectrum of CIN with HPV Status
In the below two tables/figures i.e., [Table/Fig-8,9], comparison of spectrum of CIN/SIL with p16 positivity and HR-HPV status, showed that in all studies, p16 positivity as well as HR-HPV positivity increased with the grade of dysplasia [23,24]. The mechanism of p16 overexpression is still unclear. Some researchers hypothesised that the p16 overexpression may be due to the removal of p16 inhibition by pRb, which is degraded by E7 through an ubiquitin-dependent proteinase system [25,26].
Comparison of Spectrum of CIN (CIN1,2,3) with HPV status of other studies with present study.
| CIN1 | CIN2 | CIN3 |
|---|
| p16 IHC positivity | HR-HPV positivity | p16 IHC positivity | HR-HPV positivity | p16 IHC positivity | HR-HPV positivity |
|---|
| Queiroz C et al., [8] (2006) | 53.3% | 86.7% | 70% | 100% | 93.4% | 86.6% |
| Nam EJ et al., [16] 2008) | 16.6% | 45.5% | 50% | 83.3% | 100% | 84.6% |
| Present study* (2019) | 16% | 76% | 83.4% | 91.7% | 100% | 100% |
(*Value of percentages of p16 IHC positivity are different as only those cases are taken into consideration whose HPV status was known)
Comparison of Spectrum of CIN (L-SIL and H-SIL) with HPV status of other studies with present study.
| L-SIL | H-SIL |
|---|
| p16 IHC positivity | HR-HPV positivity | p16 IHC positivity | HR-HPV positivity |
|---|
| Sano T et al., [23] (1998) | 20.6% | 70% | 83.4% | 100% |
| Lakshmi S et al., [24] (2009) | 41.2% | 0% | 87.5% | 100% |
| Present study (2019) | 16% | 76% | 87.5% | 93.8% |
*Value of percentages of p16 IHC positivity is different as only those cases are taken into consideration whose HPV status was known [23,24].
In CIN1/L-SIL cases, most studies showed a marked difference between HR-HPV and p16 positivity. This difference can be attributed to the fact that HR-HPV infection alone may not be responsible for p16 overexpression, but requires formation of aberrant E6 and E7 genes to finally overexpress p16 [25,26]. Thus, cases that were HR-HPV positive and p16 negative were cases that may not be having the aberrant genes. This suggests that p16 can be considered as a marker for carcinogenesis in cervical dysplasia. Another possibility is that the p16 negative cells are physiologically cells in a normal state. In most normal cells, p16 expression is known to be low at both the m-RNA and protein levels [27,28].
In CIN2-3/H-SIL cases, all studies showed a marked positivity in p16 IHC as well as HR-HPV levels, indicating that formation of aberrant E6 and E7 genes had been complete thereby indicating a higher grade of dysplasia.
HR-HPV positive but p16 negative CIN2 case: One CIN2 case was positive for HR-HPV but negative for p16. This may be due to the lack of formation of aberrant E6 and E7 genes or cells in a physiologically normal state [25-28].
HR-HPV and p16 negative CIN2 case: One case of CIN2 was negative for both p16 and HR-HPV. This case may be a case of pRb overexpression wherein there is an inverse relation between p16 and Rb expression, especially in HR-HPV negative CIN. Another possibility is that this case could be one of the benign mimickers of CIN like atypical immature metaplasia wherein p16 is negative and markers like P63 are positive [29]. Thus, IHC with Rb and P63 will help in sealing the diagnosis in such cases.
HPV negative but p16 positive CIN1 case: In the present study, there was only one case that was HPV negative but p16 positive. It was diagnosed as CIN1 on histopathology. A study done by Zhang G et al., [18] was performed only on HPV negative patients to check for p16 status, in which it was concluded that p16 positive cases have greater probability to progress to higher grades of dysplasia and regular follow-up of such cases is definitely required
Comparison of Prevalence of HPV Serotypes in Various Studies with Present Study
In a study of CIN biopsies, Clifford GM et al., had 45% cases positive for HPV 16, 7.1% cases positive for HPV-18 and 46.4% cases positive for Other HR-HPV [30]. Another study, done by Franceschi S et al., [31] had 22.8% cases positive for HPV 16, 4.3% cases positive for HPV-18 and 52.4% cases positive for Other HR-HPV. In the present study, 36.6% cases were positive for HPV 16, 4.9% cases were positive for HPV-18 and 63.4% cases were positive for Other HR-HPV.
Comparison of various studies with present study, in terms of prevalence of HPV serotypes, it was seen that HPV16 was the most common serotype in all studies, with HPV18, the second most common individual serotype. The ‘Other HR-HPV’ group is also significantly large in all the studies. Munoz N et al., states that a vaccine with only HPV16 and HPV18 could potentially prevent 71% of cervical cancers [32]. In contrast, a vaccine containing the 7 most common HPV types would prevent about 87% of cervical cancers worldwide. Considering the large percentage the Other HR-HPV group, it is suggested that the other less common HR-HPV serotypes also should be added to make the present vaccine (that contains only HPV 16 ad 18 among HR-HPV) to make it more effective.
Correlation of CIN Cases with p16 Expression (Cellular Pattern) and HPV Serotypes
Laxmi S et al., found that HPV16/18 positive H-SIL cases mainly showed focal to diffuse p16 IHC staining which correlated with the present study [24]. None of the cases of ‘Other HR-HPV’ showed p16IHC positivity in H-SIL cases in the study done by Laxmi S et al., but the present study proved otherwise [24]. It suggests that irrespective of serotype of High risk HPV, any serotype causing formation of aberrant E6 and E7 genes, can give rise to p16 overexpression and carcinogenesis.
To summarise, the highlights of the study were; the mean age of CIN3 cases (32.3 yr) was lower than that of CIN 1 and CIN2 cases which may be due to an unknown confounding factor or HPV16 causing direct progression to CIN3. While comparing, the percentage of p16 positive cells, the intensity of IHC reaction (weak, variable or strong), cellular reaction pattern (focal or diffused) and p16 IHC positivity in the spectrum of CIN, all these parameters increased with increasing grades of dysplasia. Due to wide variation in results of p16 IHC positivity in various studies, a scoring system similar to the one used in the present study, will definitely help to standardise results of p16 IHC. While comparing morphologically, p16 IHC intracellular pattern, weak cytoplasmic positivity of p16 was seen in lower grades of CIN while strong cytoplasmic positivity was seen in higher grades of CIN and this feature can be used as an adjunct in differentiating between low and high grades of CIN. The present study showed that in terms of differentiating between H-SIL (CIN2 and CIN3) vs L-SIL and also CIN2 vs CIN1, p16 IHC was helpful and statistically significant however p16 was not helpful or statistically significant in differentiating CIN3 vs CIN2. In the present study, p16 IHC and High risk HPV status showed strong association in H-SIL (CIN2 and CIN3) cases but no or negligible association in L-SIL (CIN1) cases. It was seen that HPV16 was the most common serotype in all studies, with HPV18, the second most common individual serotype. The ‘Other HR-HPV’ group is also significantly large in the present study.
Limitation(s)
The limitation of the study is its relative small sample size.
Conclusion(s)
Using p16 as an adjunct to histopathology will definitely help standardise reporting of grades of CIN. There is a strong association between p16 IHC and HPV-DNA in H-SIL cases. It is suggested that other less common HR- HPV serotypes also should be added to the current HPV vaccine in India, to make the present HPV vaccine (that contains only HPV 16 ad 18 among HR-HPV) more effective and probably a study with larger sample size or meta-analysis of all the p16 IHC and HPV DNA studies done in India will help consolidate its results.