The Coronary Artery Disease (CAD) is one of the most common leading cause of morbidity and mortality [1]. Apoptosis is an energy-requiring process which depends on specific signalling cascades for cell death. It regulates various physiological activities and plays a vital role in numerous pathologic conditions like cancer, autoimmune, neurological diseases and CAD [1]. Apoptotic cells are found in the various stages of coronary atherosclerotic plaques and also involves in plaque growth, lipid core development, plaque rupture and thrombosis [2].
The receptor (extrinsic) and mitochondrial (intrinsic) mediated apoptotic pathways employ the cascade of cysteine-dependent aspartate-specific proteases (Caspases). Caspases (CASP) have been broadly classified as initiator caspases (Caspase-8 and -9) and executioner caspases (Caspase-3, -6 and -7).
Cellular stress causes the release of Cytochrome-c from mitochondria leading to the activation of Caspase 9 [3]. Both apoptotic pathways transmit death signals to the effector Caspases (CASP -3, -6 and -7) to execute the events of apoptosis [4,5].
CASP9 gene mapped on chromosome 1p34-1p3.1 contains 9 exons and encodes 4.6 kDa pro-caspase proteins of 416 amino acids [6]. Single Nucleotide Polymorphisms (SNP) in genes regulating the apoptosis may affect the expression or activities of enzymes and also contribute to individual susceptibility to diseases [7]. Many studies on promoter polymorphisms of CASP9 gene demonstrated its association with various cancers [5,6]. However, the role of SNPs in promoter of CASP9 gene in CAD remains unexplored.
It has been hypothesised that promoter variants and haplotype might influence CASP9 expression, thereby modulating susceptibility to CAD. Till date there are no studies reported on specific association between CASP9 promoter polymorphisms and susceptibility to CAD and is the part of the study which has been already published [8].
Therefore, the present case-control study was performed to evaluate the association between CASP9 genotypes/haplotypes and the susceptibility to CAD in South Indian Population.
Materials and Methods
Ethical Statement
The present Case-Control study involving human participants was in accordance with the ethical standards of Institutional Ethics Committee for Biomedical Research, Institute of Genetics and Hospital for Genetic diseases IEC No. 03/IEC/IOG/OU/13, Osmania University, Hyderabad, Telangana, India and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Study Population
Criteria for inclusion and exclusion
This case-control study consisted of 300 CAD patients, age and gender matched 300 healthy controls. The sample size for the present study was calculated using online software tool Open epi version 3.0. Angiogram/clinical investigations were performed on all CAD patients admitted between January 2012 and December 2016 in the Department of Cardiology, Durgabai Deshmukh Hospital and Research Centre, Hyderabad, Telangana State, India. Patients with history of cancers, infectious diseases were excluded from this study.
Control subjects were randomly selected from a pool of healthy volunteers, who visited for the general health check-up at Durgabai Deshmukh Hospital and Research Centre, Hyderabad, Telangana State, India during the same period. Written informed consent was obtained from all the study participants.
Genotyping of CASP9 Gene Promoter Polymorphisms
5 mL of fasting peripheral venous blood was collected in EDTA and Clot activator vacutainers from all participants.
Extraction of DNA
Genomic DNA was extracted from peripheral blood leukocyte pellet using the standard salting out method [8] as shown in [Table/Fig-1]. Venous blood (5 mL) was collected in an anti-coagulant (EDTA) contained vacutainer for the extraction of genomic DNA. In the first step, equal volume of low salt buffer (TKM1) and 100 μl of Triton-X were added to lyse the red blood cells. The sample was centrifuged at 2500rpm for 10 minutes. The supernatant solution was discarded and TKM1 was added to pellet and centrifugation was repeated until the red blood cells were washed -out completely from the pellet. To the pellet, high salt buffer (TKM2), 125 μL of 10% sodiumdodecyl sulfate were added to lyse the white blood cells and 6 M sodium chloride was added to precipitate the proteins. Finally to the supernatant double volume of 100% ethanol was added to precipitate the genomic DNA.
Agarose gel image of genomic DNA (Agarose gel electrophoresis of controls and cases Lane 1-3 (CAD cases genomic DNA) and 4 to 6-controls genomic DNA).
The genotyping of three promoter SNPs of CASP9-1263G>A (rs4645978), -905T>G (rs4645980) and-712C>T (rs4645981) were carried out by PCR-RFLP.
The following primers forward: 5′-GGGAATACTTTCCTGGCAGG-3′, reverse: 5′-GTCTTC CATTCCCTCTTCCCTC-3′, forward 5′-GAA GAGGGAATGGAAGACTGTG-3′, reverse 5′-GCCCCCGGGGTCAA TCCTCA-3 and forward: 5′-AGTCGCGGAGGTGCCGCCTT-3′, reverse 5′-5-AGGGCTAGCCTCGTGCCACC -3′ and were used to amplify the 234 bp of-1263G>A, 449 bp of -905T>G and 194 bp of -712C>T, respectively. The PCR reactions was performed in total volume of 10 μL containing 50 ng genomic DNA, 10 pmol/μL of each primer, 2.5 mM of each dNTP, 10X PCR buffer, 25 mM MgCl2, 5U/μL of Taq polymerase. The amplication conditions include denaturation at 95°C at 60 seconds and annealing termperatures at 58.7°C (-1263G>A), 55.2°C (-905T>G) and 60.0°C (-712C>T) for 35 seconds respectively and extension at 72°C for 35 seconds and the same was repeated for 35 cycles. The PCR products were visualised on 3% agarose gel with ethidium bromide staining. PCR products of CASP9-1263G>A, -905T>G and -712C>T were digested with 1U of BsmAI, HaeII and BseY1 restriction enzymes (NEB) as per the manufacturer instructions.
Statistical Analysis
Demographic and clinical characteristics of patients and controls are presented as mean and standard deviation. The data of CAD cases and controls were compared using the Student’s t-test for continuous variables and a Chi-square test for categorical variables by using online statistical tools such as Interactive Chi-square analysis and OpenEpi version 3.03. Deviations of the genotype frequencies were assessed by Hardy-Weinberg calculator [9]. Risk estimates were calculated for co-dominant, dominant and recessive genetic models using SNP Stats [10]. Haplotype analysis and LD between the promoter SNPs of the CASP9 gene were analysed by Haploview software (v4.2) [11]. Promoter SNPs of CASP9 and their interactions with epidemiological factors were characterised by MDR [12,13].
Results
In the present study, the mean age of control and CAD patients were 54.55±14.30 years and 55.22±13.61 years respectively. The demographic and clinical characteristics of the cases and controls were compared and found that the variables such as BMI, Blood pressure, Smoking, Alcohol and lipid profiles showed significant difference (p<0.05) between CAD patients and controls and the details have been published in our earlier study [8]. Range of lipid profile was higher in patients which included levels of total cholesterol, triglyceride, LDL, and mean values of these lipid markers (mg/dL) in CAD patients were 265.28±41.56, 181.11±51.59, 145.47±41.7 respectively while HDL cholesterol was lower in patients with mean value 30.73±9.12 compared to controls.
Molecular Analysis
Allelic, genotypic distributions and risk genotypes of CASP 9-1263G>A (rs4645978), -905T>G (rs4645980) and -712C>T (rs4645981) promoter polymorphisms were shown in [Table/Fig-2,3]. The allelic frequencies of these three promoter SNPs of CASP 9 in controls were consistent with Hardy-Weinberg Equilibrium.
Allele and genotypic distribution of caspase 9 gene promoter polymorphisms (-1263G>A, -905T>G and -712C>T) in controls and CAD patients.
| Gene | Marker | Controls (n=300) | CAD patients (n=300) | Chi-square | p-value | Allele frequencies | HWE p-value |
|---|
| Controls | CAD patients |
|---|
| MM | Mm | mm | MM | Mm | mm | | | M allele | m allele | M allele | m allele | Controls | CAD patients |
|---|
| CASP9 | -1263G>A | 74(24.7%) | 135(45%) | 91(30.3%) | 127(42.3%) | 81(27%) | 92(30.7%) | 27.48 | <0.05* | 0.47 | 0.53 | 0.56 | 0.44 | 0.1 | <0.0001* |
| -905T>G | 125(41.7%) | 134(44.7%) | 41(13.7%) | 61(20.3%) | 167(56%) | 72(23.7%) | 33.88 | <0.05* | 0.64 | 0.36 | 0.48 | 0.52 | 0.62 | 0.049 |
| -712C>T | 180(60%) | 99(33%) | 21(7%) | 32(10.3%) | 197(66%) | 71(23%) | 165.39 | <0.05* | 0.76 | 0.24 | 0.43 | 0.57 | 0.15 | <0.0001* |
M: Major allele; m: Minor allele; HWE: Hardy-weinberg equilibrium; *statistically significant (p-value <0.05) (Interactive Chi-square analysis and Hardy-Weinberg calculator, SNP STATS)
Risk estimates of Caspase 9 promoter polymorphisms (-1263G>A, -905T>G & -712C>T) in controls and CAD patients (n=600).
| Model | CASP9 -1263G>A | CASP9 -905T>G | CASP9 -712C>T |
|---|
| Genotype | Odds ratio | p-value | Genotype | Odds ratio | p-value | Genotype | Odds ratio | p-value |
|---|
| Co-dominant | GG | 1.00 | | TT | 1.00 | | CC | 1.00 | <0.05* |
| GA | 0.25 (0.12-0.52) | <0.05* | TG | 3.51 (1.69-7.29) | <0.05* | CT | 13.36 (6.40-27.91) | <0.05* |
| AA | 0.39 (0.18-0.84) | <0.05* | GG | 5.61 (2.21-14.24) | 0.15 | TT | 17.64 (6.38-48.79) | <0.05* |
| Dominant | GG | 1.00 | | TT | 1.00 | | CC | 1.00 | <0.05* |
| GA-AA | 0.30 (0.15-0.60) | <0.05* | TG-GG | 3.88 (1.89-7.94) | <0.05* | CT-TT | 14.19 (6.96-28.94) | |
| Recessive | GG-GA | 1.00 | | TT-TG | 1.00 | | CC-CT | 1.00 | 0.0034* |
| AA | 0.93 (0.52-1.66) | 0.8 | GG | 2.08 (1.01-4.26) | 0.041* | TT | 3.35 (1.41-7.95) | |
| Over dominant | GG-AA | 1.00 | | TT-GG | 1.00 | | CC-TT | 1.00 | <0.05* |
| GA | 0.43 (0.24-0.75) | 0.002* | TG | 1.48 (0.85-2.55) | 0.16 | CT | 4.77 (2.67-8.51) | <0.05* |
*statistically significant (p-value <0.05) (SNP STATS)
Molecular analysis revealed that the patients with TT genotype of CASP9 -712 C>T polymorphism showed 17.64 fold risk and TG genotype of CASP9 -905 T>G polymorphism have 3.51 fold risk for development of CAD while GA and AA genotypes of CASP9 -1263 G>A were significantly conferring protective effect on CAD.
Transcription Factor Binding Site analysis (TFBS) by Alibaba online tool (version 2.1) (http://gene-regulation.com/pub/programs/alibaba2/index.html).
TFBS revealed that CASP9 -1263G>A polymorphism with G allele has 3 Sp1 transcription factor binding sites while variant A allele has only 2 Sp1 transcription factor binding sites, CASP9 -905T>G polymorphism with T allele have Sp1-Nf-muF1 binding sites where as G allele have Nf-muF1-Sp1 transcription binding sites and CASP9 -712C>T polymorphism with T allele has Sp1 transcription factor binding site while C allele have krox-20, Sp1, NF-1, ETF binding sites.
Linkage Disequilibrium
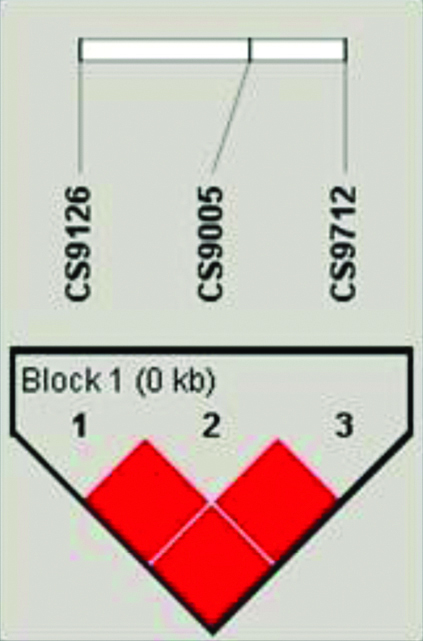
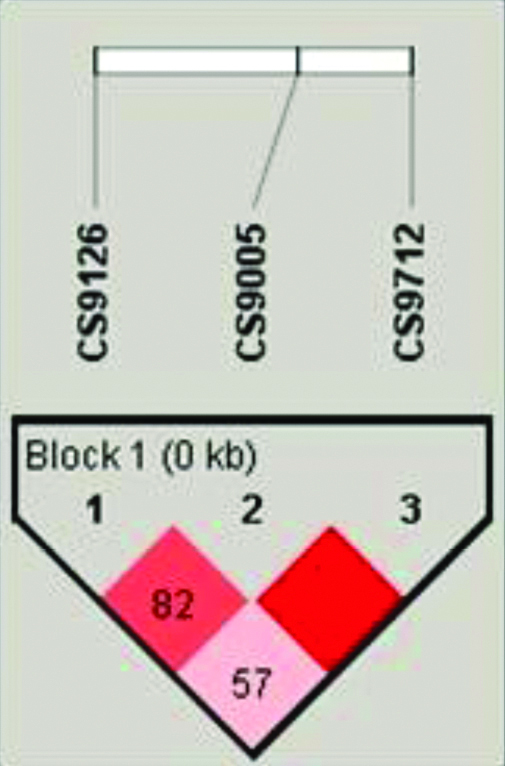
Measuring the levels of LD is important for gene mapping of complex disease loci and understanding of genome architecture. In the present study, LD coefficients and haplotype block structure were calculated using Haploview (version 4.1) software. In the present study, strong LD (D′=1) was observed between all the SNPs of Caspase 9 promoter polymorphisms in controls, while CASP9 -905T>G, CASP9-712C>T showed perfect LD in CAD patients as shown in [Table/Fig-4,5].
LD plot of CASP9 promoter polymorphisms (-1263G>A, -905T>G and -712C>T) in controls.
LD plot of CASP9 promoter polymorphisms (-1263G>A, -905T>G and -712C>T) in CAD patients.
Haplotype Analysis
Out of 6 haplotypes of CASP9 promoter polymorphisms, G-T-C (0.457:0.188 χ2=99.59, p<0.05), A-T-C (0.172:0.044 χ2=50.50, p<0.05), A-G-C (0.136: 0.201, χ2=9.034, p=0.002), G-T-T (0.011: 0.251, χ2= 151.63, p<0.05), G-G-T (0.003: 0.119, χ2=70.103, p<0.05) haplotypes of -1263G>A, -905T>G, -712C>T polymorphisms conferred risk with a significant case control ratio for developing CAD as shown in [Table/Fig-6].
Haplotype combinations of caspase 9 promoter polymorphisms (-1263G>A, -905T>G and -712C>T).
| Haplotype | Frequency | Case: Control ratios | Chi-square | p-value |
|---|
| Haplotype combinations of Caspase 9 promoter (-1263, -905, -712) polymorphisms |
| G-T-C | 0.323 | 0.457:0.188* | 99.59 | <0.05* |
| A-G-T | 0.208 | 0.221:0.196 | 1.073 | 0.300 |
| A-G-C | 0.169 | 0.136:0.201 | 9.034 | 0.002* |
| G-T-T | 0.131 | 0.011:0.251 | 151.63 | <0.05* |
| A-T-C | 0.108 | 0.172:0.044* | 50.50 | <0.05* |
| G-G-T | 0.061 | 0.003:0.119 | 70.103 | <0.05* |
*Significant case: control ratio (p-value <0.05) chi-square test Haploview (version 4.1) software)
Multifactor Dimensionality Reduction (MDR) analysis
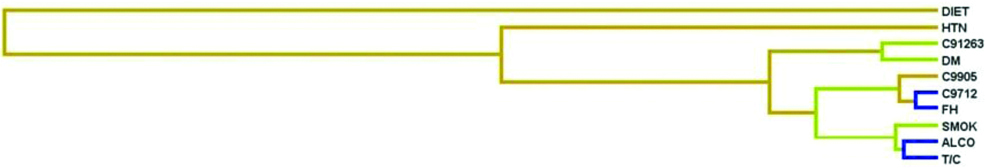
The interaction analysis between promoter SNPs of CASP9 gene and epidemiological factors is shown in dendrogram [Table/Fig-7]. Interaction between SNPs and epidemiological parameters showed that CASP9-712C>T polymorphism with positive family history, consumption of beverages (tea/coffee) and alcohol have high degree of redundancy, CASP9 -1263A>G polymorphism and DM, smoking have reduced degree of redundancy and other markers CASP9 -905T>G, polymorphism, diet, hypertension have independent effect on the disease phenotype. The results of the MDR analysis were evaluated for possible combinations of Caspase 9 promoter polymorphisms for the risk of developing CAD as summarised in [Table/Fig-8]. It was observed that combination of GG genotype of Caspase -905T>G, AA genotype of -1263G>A, CT genotype of -712C>T promoter polymorphism was found to be the best model along with its least prediction error of 0.112 and CV consistency of 10/10 conferring a 2 fold risk (40/20) for the occurrence of apoptotic events in CAD as shown in [Table/Fig-9].
Dendrogram: CASP9 promoter polymorphisms (-1263G>A, -905T>G and -712C>T) and epidemiological factors interactions.
MDR-best model for CASP9 promoter polymorphisms (-1263G>A, -905T>G and -712C>T).
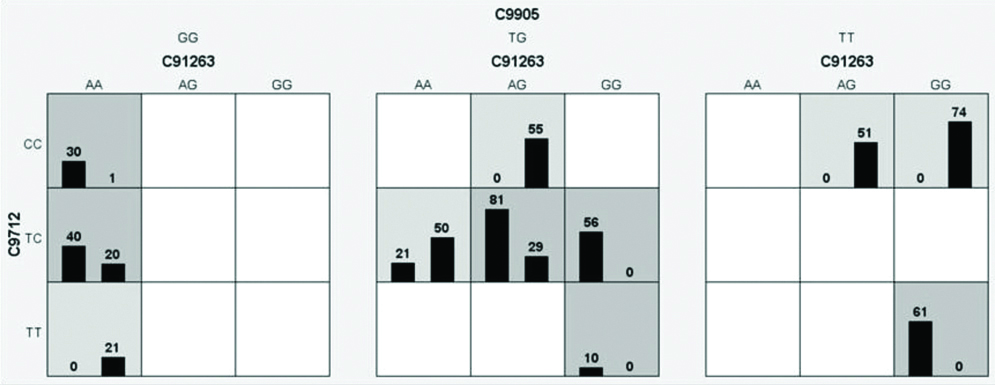
MDR-best model for CASP9 promoter polymorphisms (-1263G>A, -905T>G and -712C>T).
| Caspase 9 promoter polymorphisms | Trainining BalAcc | Testing BalAcc | Testing error | CV consistency |
|---|
| Caspase 9 -712C>T | 0.7505 | 0.7505 | 0.2495 | 10/10 |
| Caspase 9 -1263G>A, -712C>T | 0.8482 | 0.8482 | 0.1518 | 10/10 |
| Caspase 9 -1263G>A, -712C>T, -905T>G | 0.8818 | 0.8818 | 0.1182* | 10/10 |
*Least testing error
Discussion
Apoptosis is associated with the integrity of vascular smooth muscle cells and stability of atherosclerotic plaque in coronary arteries [14]. Apoptosis is genetically determined and the functional polymorphisms in its genes regulate the extrinsic and intrinsic pathways. SNPs in Caspase genes may modulate the apoptosis and contribute to the variations in the phenotype [8,15]. Therefore, we have studied the promoter polymorphisms of Caspase-9 (-1263G>A, -905T>G, -712C>T) in CAD cases and controls and we have found that -1263 GA and AA genotypes conferred significant protective effect for CAD. Conversely, Park JY et al., found that the -1263 GG genotypes was associated with a significantly decreased risk of lung cancer compared with the -1263 AA genotype in the Korean population [15].
TFBS analysis showed that the -1263 A allele has 2 Sp1 transcription factor binding sites whereas G allele has 3 Sp1 binding sites, the difference in the number of Sp1 binding sites might alter the transcriptional activity of the gene. Another SNP at promoter region -712C>T was studied by Park JY et al., and found that the individuals with at least one -712 T allele were at a significantly increased risk of lung cancer compared with those harbouring the -712 CC genotype [15]. A study by Theodoropoulos GE et al., also found that T allele and TT genotype were significantly associated with the increased risk of breast cancer compared with the CC genotype [16]. Similarly the present study also revealed that the ‘TT’ genotype is significantly associated with CAD. An analysis of potential transcription factor-binding sites of -712C>T polymorphism revealed that C allele with Sp1 and krox-20, NF-1, ETF binding sites may regulate the transcription, while T allele with only Sp1 transcription factor binding site might increase the transcriptional activity of the gene.
Further, a third polymorphism-905T>G of the Caspase 9 gene was studied by Park JY et al., and did not find any association with the risk of lung cancer [15]. In the present study, we have found that TG genotype and G allele are strongly associated with risk of CAD. An analysis of the potential transcription factor-binding sites showed that the -905 G allele have Nf-muF1-Sp1-binding sites, whereas the T allele has Sp1-Nf-muF1 transcription factor binding sites. The change in position of Sp1 binding sites might cause the alterations in the promoter activity of the gene. Haplotype analysis is useful to investigate the impact of the group of alleles on risk of disease. To know the effect of three CASP9 polymorphisms (-1263G>A, -905T>G and -712 C>T) we have performed haplotype analysis and found that G-T-C (0.457:0.188 χ2=99.59, p<0.05) and A-T-C (0.172:0.044 χ2=50.50, p<0.05) haplotypes conferred risk with a significant case, control ratio for developing CAD.
MDR analysis for gene-epidemiological interactions revealed that interaction between SNPs and epidemiological parameters showed that CASP9 -712C>T polymorphism, family history, consumption of beverages (tea/coffee) and alcohol have high degree of redundancy, CASP9 -1263A>G polymorphism and DM, smoking has reduced degree of redundancy and other markers CASP9 -905T>G, polymorphism, diet, hypertension have an independent or additive effect on the disease phenotype.
Limitation(s)
Present study has limitations having a small sample size and the study subjects were restricted to the South India. Polymorphisms vary with ethnically diverse populations. The mechanisms underlying the associations of caspase 9 gene polymorphism with the risk of CAD are necessary to emphasise with the help of collaborative studies on different populations.
Conclusion(s)
Present study implicated that CASP9 -905 TG and -712 TT polymorphic variants and G-T-C, A-T-C haplotype might be associated with susceptibility to CAD. Apoptosis, a genetically programmed cell death is involved in various physiological and pathological events. The regulation of apoptosis is emerging as a potential therapeutic strategy for CAD. Identifying a molecular apoptotic marker may guide to detect the high risk individuals and offer new therapeutic strategies to improve diagnosis and prognosis of CAD patients.