Antimicrobial resistance is a public health concern that requires a global coordinated action with a view to address its rising threat. Antimicrobial resistance has been increasing in prevalence worldwide in both developed and developing countries [1,2]. Most worrisome is limited resources in developing countries, which has inadvertently exacerbated the growing threat of antimicrobial resistance [2]. The annual death resulting from antibiotic resistant infections has been estimated globally at about 700,000 people and the deaths have been projected to hit 10 million by 2050 [3]. About half of those deaths are projected to occur in Africa because of non-effectiveness of currently available antibiotics and the non-availability and non-affordability of highly potent or alternative antibiotics [3].
Beta-lactamase production by Enterobacteriaceae is the most important single mechanism of resistance to beta-lactam antibiotics (penicillins, cephalosporins, monobactams and carbapenems), which are often used to treat nosocomial and community acquired infections. ESBL enzymes which are commonly produced by E. coli and Klebsiella pneumoniae hydrolyze third generation cephalosporins and monobactam thus rendering them ineffective against ESBL producing organisms, thus increasing costs, length of hospital stay, burden of disease and ultimately morbidity and mortality rates [4]. ESBL hydrolyze beta-lactams such as cefotaxime, ceftriaxone and ceftazidime, ampicillin, penicillins and aztreonam [5].
Materials and Methods
This was a prospective convenient cross-sectional study. Four hundred and ninety-eight clinical specimens were obtained using formulae, N=Z2Pq/d2 [12] based on the prevalence of 79.8% [13]. The specimens (included ear swab, wound, eye swab, urine, stool, high vaginal swab, cerebrospinal fluid and blood culture) were collected between November 2017 and November 2018 at the Aminu Kano Teaching Hospital, Kano, Federal Medical Centre, Katsina and Asokoro District Hospital, Abuja. Both outpatients and inpatients of these hospitals were recruited into the study following ethical approvals and informed consent from the patients. Ethical approvals were sought and obtained from the Research and Ethics Committee of the three hospitals with reference numbers; FHREC/2018/01/95/14-08-18 for Asokoro District Hospital, Abuja, NHREC/21/08/2008/AKTH/EC/2301 for Aminu Kano Teaching Hospital, Kano, while Federal Medical Centre, Katsina had no reference number but approval letter was obtained.
The patients recruited into the study had age range of <11 months to 71 years with both males and females. Clinical and demographic data, such as age, gender and patient status (Inpatient or Outpatient) were collected from all patients using a proforma.
Inclusion criteria: All patients that endorsed informed consent form and filled questionnaire (questions such as age, sex, recent use of antibiotics, whether it was self-medication or by prescription, history of travel and so on) were included in the study from the three hospitals.
Exclusion criteria: All patients that refused to sign or thumb print the informed consent forms and questionnaires were excluded from the study.
AKTH and FMCK are tertiary hospitals while ADHA is a secondary hospital.
Cultural Isolation and Identification
Clinical samples were cultured on Blood and MacConkey agars (Oxoid, U.K) overnight at 37°C and E. coli was identified using conventional microbiological procedures [14]. All E. coli isolates were further confirmed using Microbact 12A/12E (Oxoid, UK). This identification and other experiments were carried out at Molecular Biology laboratory of Department of Medical Laboratory Science, Ladoke Akintola University of Technology, Osogbo, Nigeria.
Antimicrobial Susceptibility Testing
The antibiotic susceptibility patterns of the 104 E. coli isolates to a panel of 9 antibiotics including representative of third generation cephalosporin, comprising ceftazidime (30 μg), cefotaxime (30 μg), meropenem (10 μg), imipenem (10 μg), ciprofloxacin (5 μg), gentamicin (10 μg), ertapenem (10 μg), amoxy-clavulanate (30 μg), Ampicillin (10 μg) were determined by the disc diffusion method (Oxoid, UK) using Mueller-Hinton agar (Oxoid, UK) according to CLSI guidelines [15]. Isolates were further selected for minimum inhibitory concentrations to ceftazidime, amoxyclav, ciprofloxacin, ampicillin, cefotaxime and gentamicin using agar dilution method. All runs included the control organism E. coli ATCC 25922.
Detection and Identification of Beta-lactamase Genes
All isolates were tested for production of ESBL/AmpC and carbapenemase based on susceptibility testing using the disc based ‘ESBL/AmpC and Carbapenemase detection set’ from Mast Diagnostics (Bootle, UK) following manufacturer’s protocol and interpreted using the ‘ESBL/AmpC and carbapenemase detection set calculator’ tools also as per the manufacturer’s guidelines. Polymerase Chain Reaction (PCR) was used to detect β-lactam resistance genes from 44 isolates (selected to cover all phenotypes present) using primers shown in [Table/Fig-1] as previously described [7,16,17]. Control strains obtained from the Health Protection Agency (HPA) (now part of Public Health England, PHE) included for each gene. Amplimers resulting from these PCR reactions were sequenced to confirm the identity and specific variant of each gene identified and sequences were aligned to known reference sequences using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo).
Primers used for amplification of β-lactamase genes.
| Primer | DNA sequence (5′ to 3′) | Annealing temp (°C) | Product size (bp) |
|---|
| NDM FNDM R | TTGATGCTGAGCGGGTGCTGTCCTTGATCAGGCAGC | 56 | 578 |
| KPC FKPC R | ATGTCACTGTATCGCCGTCTTAGACGGCCAACACAATAGG | 56 | 785 |
| OXA 48 FOXA 48 R | TTCGGCCACGGAGCAAATCAGGATGTGGGCATATCCATATTCATCGCA | 56 | 240 |
| VIM FVIM R | AGTGGTGAGTATCCGACAGATGAAAGTGCGTGGAGAC | 56 | 261 |
| IMP FIMP R | CTACCGCAGCAGAGTCTTTGAACCAGTTTTGCCTTACCAT | 58 | 587 |
| GES FGES R | CGGTTTCTAGCATCGGGACACATCCGCCATAGAGGACTTTAGCACAG | 58 | 263 |
| SME FSME R | AACGGCTTCATTTTTGTTTAGGCTTCCGCAATAGTTTTATCA | 58 | 830 |
| IMI FIMI R | GAGGGTATGACTAAATTCATGCGGTCGAGCAGGTGTAGATGTGTCACGTCATCG | 58 | 116 |
| PER-1FPER-1R | ATGAATGTCATTATAAAAGCAATTTGGGCTTAGGGCAGAA | 51 | 590 |
| VEB FVEB R | CGACTTCCATTTCCCGATGCGGACTCTGCACCAAATACGC | 55 | 604 |
| OXA-10 FOXA-10 R | GTCTTTCGAGTACGGCATTAATTTTCTTAGCGGCAACTTAC | 52 | 600 |
| OXA FOXA R | ATATCTCTACTGTTGCATCTCCAAACCCTTCAAACCATCC | 50 | 216 |
| CTX-M-1 FCTX-M-1 R | GACGATGTCACTGGCTGAGCAGCCGCCGACGCTAATACA | 60 | 499 |
NDM: New Delhi Metalobeta-lactamase; KPC: Klebsiella pneumoniae carbapenemase; OXA: Oxacillinase; VIM: Verona Integron-encoded Metalobetalactamase; GES: Guiana extended spectrum beta-lactamase; PER: Pseudomonas extended spectrum resistant; VEB: Vietnam extended spectrum beta-lactamase; CTX-M: Cefotaximase munich; SME: Serratia marcescens enzyme; IMI: Imipenem hydrolysing beta-lactamase; IMP: Imipenemase
Random Amplified Polymorphic DNA and Polymerase Chain Reaction Typing
The epidemiological relationships between strains of E. coli analysed by random amplified polymorphic DNA. The primers sequence and PCR running conditions used were according to Vogel I et al., [18], modified to use 1 μL of 100 μm of primers at a final concentration of 0.02 μm [19]. The experiment was repeated to ensure reproducibility. DNA fingerprints were compared by visual inspection to assign similar banding patterns to the same Random Amplified Polymorphic DNA (RAPD) type.
Statistical Analysis
Data from this study were reported in frequency tables and percentages. The data were analysed with the aid of statistical package for social sciences (IBM SPSS), version 21.0.
Results
In total, 104 (60.5%) non-duplicate clinical E. coli strains were obtained from 172 Gram negative bacilli identified in the three hospitals (from 498 clinical specimens). Out of these 104 isolates, 61 (58.7%) were from females while 43 (41.3%) were from males. The age range of the participants in this study was between 11 months and 71 years, 70.2% was found between age range 11-40 years. Similarly, 51 (49.0%%) E. coli were found in Inpatients and 53 (51%) Outpatients. Disc susceptibility of 104 E.coli isolates showed that 93 were largely resistant to ampicillin (89.4%), 75 amoxicillin-clavulanic acid (72.1%), while ciprofloxacin and gentamicin had resistance rate of 73 (70.2%) and 64 (61.5%) respectively. More than half of E. coli were resistant to third generation cephalosporin class of antibiotics used in this study, they comprised cefotaxime with resistant rate of 74 (71.2%) and ceftazidime 53 (51.0%). Carbapenem resistance was less common; ertapenem (7.7%), imipenem (4.8%) and meropenem (2.9%) [Table/Fig-2]. MIC of six antibiotics further confirmed resistance shown by disc susceptibility with the majority of isolates having an MIC 256 μg/mL with no difference between MIC50 and MIC90 for almost all the isolates [Table/Fig-3].
Summary of antimicrobial disc susceptibility testing of 104 E. coli isolates.
| Antibiotic (μg/mL) | Sensitive n (%) | Intermediate n (%) | Resistant |
|---|
| Ceftazidime (30) | 47 (45.2) | 4 (3.8) | 53 (51.0) |
| Meropenem (10) | 101 (97.1) | 0 (0.0) | 3 (2.9) |
| Imipenem (10) | 98 (94.2) | 1 (1.0) | 5 (4.8) |
| Cefotaxime (30) | 30 (28.8) | 0 (0.0) | 74 (71.2) |
| Ciprofloxacin (5) | 26 (25.0) | 5 (4.8) | 73 (70.2) |
| Gentamicin (10) | 31 (29.8) | 9 (8.7) | 64 (61.5) |
| Ertapenem (10) | 92 (88.5) | 4 (3.8) | 8 (7.7) |
| Amoxy-clavulanate (30) | 22 (21.2) | 7 (6.7) | 75 (72.1) |
| Ampicillin (10) | 7 (6.7) | 4 (3.9) | 93 (89.4) |
Numbers in parentheses are percentages
Minimum inhibitory concentrations of E. coli isolates.
| Isolate | Antimicrobial agents | MIC (0.06 -256) μg/mL) | Sensitive n (%) | Intermediate n (%) | Resistant n (%) |
|---|
| MIC50 | MIC90 |
|---|
| Escherichia coli | Cefotaxime | 256 | 256 | 0 (0.0) | 2 (1.9) | 102 (98.1) |
| Ceftazidime | 128 | 256 | 10 (9.6) | 15 (14.4) | 79 (76.0) |
| Amoxyclav | 256 | 256 | 0 (0.0) | 3 (2.9) | 101 (97.1) |
| Ampicillin | 256 | 256 | 0 (0.0) | 0 (0.0) | 104 (100.0) |
| Ciprofloxacin | 256 | 256 | 2 (1.9) | 0 (0.0) | 102 (98.1) |
| Gentamicin | 128 | 256 | 17 (16.3) | 4 (3.9) | 83 (79.8) |
Numbers in parentheses are percentages
Phenotypic Detection of Beta-lactamases
Phenotypic detection of beta-lactamases revealed 44 isolates (42.3%) to be ESBL-producers, AmpC enzymes were present in 15 isolates (14.4%) while 27 (26.0%) were predicted to be carbapenemase producers. Analyses of sources revealed that 28 out of 51 isolates (54.9%) from inpatients produced ESBLs while for outpatients only 16 of 53 (30.2%). Breakdown according to the hospital showed that E. coli from AKTH had more ESBL and carbapenemase resistance (59.3% and 40.7% respectively) while FMCK had more putative AmpC producers (19%) than others [Table/Fig-4].
Phenotypic distribution of beta-lactamases in 104 E. coli.
| Status | Number | ESBL present n (%) | ESBL absent n (%) | AmpC present n (%) | AmpC absent n (%) | Carba present n (%) | Carba absent n (%) |
|---|
| Inpatients | 51 | 28 (54.9) | 23 (45.1) | 10 (19.6) | 41 (80.4) | 20 (39.2) | 31 (60.8) |
| Outpatient | 53 | 16 (30.2) | 37 (69.8) | 5 (9.4) | 48 (90.6) | 7 (13.2) | 46 (86.8) |
| Total | 104 | 44 (42.3) | 60 (57.7) | 15 (14.4) | 89 (85.6) | 27 (26.0) | 77 (74.0) |
| Hospital |
| FMCK | 21 | 9 (42.9) | 12 (57.1) | 4 (19.0) | 17 (81.0) | 4 (19.0) | 17 (80.9) |
| ADHA | 56 | 19 (33.9) | 37 (66.1) | 6 (10.7) | 50 (89.3) | 12 (21.4) | 44 (78.6) |
| AKTH | 27 | 16 (59.3) | 11 (40.7) | 5 (18.5) | 22 (81.5) | 11 (40.7) | 16 (59.3) |
| Total | 104 | 44 (42.3) | 60 (57.7) | 15 (14.4) | 89 (85.6) | 27 (26.0) | 77 (74.0) |
Carba: Carbapenemase. Numbers in parentheses show percentages
Genotypic Detection of ESBL
PCR and sequencing identified different ESBL encoding genes in the 44 ESBL producers. blaCTX-M were identified from 33/44 (75%) isolates, of which blaCTX-M-15 has the dominant variant for 24/33 (72.7%) isolates, all isolates had multiple ESBL genes between 2 and 5. blaCTX-M-2 had 18/33 (54.5%), blaCTX-M-9 11/33 (33.3%), blaCTX-M-8 10/33 (30.3%) and blaCTX-25 6/33 (18.2%). Other ESBL genes identified were blaOXA variants 17/44 (38.6%), of these blaOXA-10 was the dominant variant with 6/17 (35.3%), blaSHV genes were seen in 11/44 (25.0%); blaVEB from 21/44 (47.7%) and blaPER in 6/44 (13.6%) [Table/Fig-5]. However, no AmpC or carbapenemase genes were detected. ESBL genes were detected in different proportions with varying diagnosis and sources in these hospitals [Table/Fig-6]. Percentage distribution of the ESBL genes between inpatients and outpatients showed blaCTX-M/blaCTX-M-15 to be more prevalent 93.8%/56.3% in outpatients compared to inpatients 89.3%/53.6%. Among the specimens, urine exhibited highest prevalence of ESBL genes 19/44 (43.2%) followed by stool and wound having 8/44 (18.2%) and 6/44 (13.6%) respectively, least prevalence was found in Semen, Ear and Eye swabs with prevalence of 2.3% each.
Carriage of beta-lactamases in E. coli isolates.
| Serial # | ID no. | Diagnosis | Specimen | Source | MIC (μg/mL) | Phenotype | β-lactam gene |
|---|
| | | | | CTX | CFZ | AMC | Amp | CIP | GEN | E | A | C | |
|---|
| 1 | AD 03 | UTI | Urine | OP | 256 | 128 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-15, CTX-M-2 |
| 2 | AD 04 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | PER, VEB, SHV, OXA |
| 3 | AD 05 | GET | Stool | IP | 256 | 128 | 128 | 256 | 128 | 256 | + | + | - | CTX-M-15, CTX-M-9 |
| 4 | AD 06 | GET | Stool | OP | 256 | 256 | 256 | 256 | 256 | 128 | + | - | - | CTX-M-15, CTX-M-2, VEB |
| 5 | AD 07 | Sepsis | Wound | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | SHV, CTX-M-15, OXA |
| 6 | AD 08 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | + | - | CTX-M-2, CTXM-8, CTX-M-15, |
| 7 | AD 09 | GET | Stool | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, OXA, CTX-M-2, VEB |
| 8 | AD 17 | GTI | HVS | OP | 256 | 256 | 256 | 256 | 128 | 256 | + | + | + | CTXM-8, VEB, CTX-M-15 |
| 9 | AD 21 | Sepsis | Wound | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-15, OXA, CTX-M-25 |
| 10 | AD 22 | UTI | Urine | IP | 256 | 128 | 256 | 256 | 256 | 256 | + | - | - | VEB, CTX-M-15 |
| 11 | AD 26 | Sepsis | Wound | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M, SHV, OXA-10, CTX-M-2 |
| 12 | AD 30 | GTI | HVS | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | + | - | CTX-M-2 VEB, |
| 13 | AD 47 | Infertility | Semen | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | SHV, CTX-M-15 |
| 14 | AD 52 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-15, CTX-M-2, |
| 15 | AD 56 | GTI | HVS | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M, VEB, |
| 16 | AD 60 | UTI | Urine | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | + | - | PER, VEB, CTX-M-15 |
| 17 | AD 65 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 128 | + | - | + | CTX-M, OXA, VEB, OXA-10 |
| 18 | AD 78 | GTI | HVS | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | SHV, VEB, CTX-M-2 |
| 19 | AD 84 | UTI | Urine | OP | 256 | 256 | 256 | 256 | 128 | 256 | + | + | - | VEB, SHV, CTX-M-8 |
| 20 | AK 08 | Septicaemia | Blood | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, VEB, SHV, CTX-M-9 |
| 21 | AK 11 | UTI | Urine | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-8, CTX-M-25, VEB, CTX-M-15 |
| 22 | AK 16 | GET | Stool | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-15, CTX-M-25, PER |
| 23 | AK 17 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 128 | + | - | + | CTX-M-15, CTX-M-9, VEB, OXA |
| 24 | AK 20 | Sepsis | Wound | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, CTX-M-2, CTX-M-9, |
| 25 | AK 22 | GTI | HVS | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | SHV, CTX-M-8 CTX-M-2, OXA |
| 26 | AK 23 | GTI | HVS | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, CTX-M-8, OXA, OXA-10 |
| 27 | AK 24 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 128 | 256 | + | - | + | CTX-M-9, CTX-M-2, SHV, |
| 28 | AK 27 | UTI | Urine | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-9, VEB, CTX-M-2 |
| 29 | AK 28 | GET | Stool | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-2, VEB, OXA |
| 30 | AK 31 | Septicaemia | Blood | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15 SHV, CTX-M-8, VEB |
| 31 | AK 32 | Otitis media | Ear swab | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, VEB, CTX-M-25 |
| 32 | AK 41 | Conjunctivitis | Eye swab | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-2, CTX-M-9, OXA, PER |
| 33 | AK 49 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-8, CTX-M-2, CTX-25 |
| 34 | AK 53 | UTI | Urine | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | + | + | OXA, OXA-10, PER |
| 35 | AK 59 | Sepsis | Wound | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | OXA, CTX-M-2, CTX-M-9 |
| 36 | FM 06 | UTI | Urine | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | + | - | CTX-M-15, CTX-M-25, OXA |
| 37 | FM 10 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 128 | + | - | - | CTX-M-2 SHV, VEB, CTX-M-9 |
| 38 | FM 14 | GET | Stool | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | OXA, SHV, CTX-M-8 |
| 39 | FM 15 | Sepsis | Wound | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, VEB, CTX-M-9 |
| 40 | FM 19 | GET | Stool | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | OXA, OXA-10, PER |
| 41 | FM 21 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-8, CTX-M-15, CTX-M-2 |
| 42 | FM 25 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | - | CTX-M-15, SHV, CTX-M-9, OXA |
| 43 | FM 31 | GET | Stool | OP | 256 | 256 | 256 | 256 | 256 | 256 | + | - | + | CTX-M-8, CTX-M-2 |
| 44 | FM 36 | UTI | Urine | IP | 256 | 256 | 256 | 256 | 256 | 256 | + | + | - | VEB, OXA, OXA-10 |
AD: Asokoro District Hospital Abuja; AK: Aminu Kano Teaching Hospital, Kano; FM: Federal medical centre, Katsina; CTX: Cefotaxime; CFZ: Ceftazidime; AMC: Amoxycillin-clavulanic acid; Amp: Ampicillin; CIP: Ciprofloxacin; GEN: Gentamicin; UTI: Urinary tract infection; GET: Gastroenteritis; GTI: Genital tract infection; HVS: High vaginal swab; OP: Outpatient; IP: Inpatient; E: Extended spectrum β-lactamase; A: AmpC; C: Carbapenemases; OXA: Oxacillinase; PER: Pseudomonas extended spectrum resistant; VEB: Vietnam extended spectrum beta-lactamase; CTX-M: Cefotaximase munich
Distribution of ESBL genes between inpatients and outpatients.
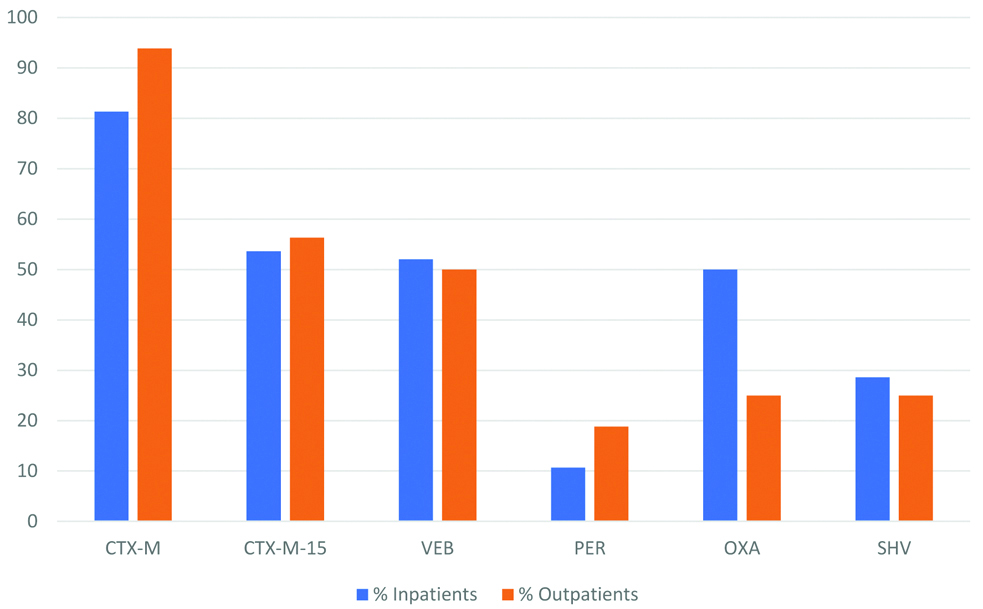
Typing of Isolates
RAPD typing was used to determine the degree of clonality among the ESBL producing E. coli isolates. The data revealed high diversity amongst all the species tested, with no identical RAPD patterns observed.
This suggests that the spread of resistance genes is underpinning the spread of resistance rather than expansion of a dominant clones.
Discussion
It is a fact that ESBLs production is the most common mechanism of E. coli resistance to third-generation cephalosporins among Enterobacteriaceae and it mediates multidrug resistance, as shown in this study. Phenotypic production of ESBLs was seen in 42.3% of isolates tested which were not closely related. This was considerably higher than other reports from similar studies from different parts of the world including different cities/regions of Nigeria where prevalences of ESBL production between 18.6% and 42% had been reported in Nigeria [8,10,11]. While outside Nigeria; in Benin, Tanzania, Mexico, Nepal and India ESBL report was between 31.3 and 38.7% [20-24]. But in Turkey and USA, ESBL productions were extremely higher 84.0% and 72% respectively [25,26]. The differences in prevalence may be multifaceted ranging from varying sample size, ESBL detection method, population dynamics, literacy, economic, socio-cultural or lack or inadequate antimicrobial stewardship. The presence of ESBL-producers in an individual is a key indicator to increased antibiotic resistance because plasmid carries multiple resistance genes including ESBL.
The prevalence of ESBL production in both inpatients and outpatients were 54.9% and 30.2%, respectively. This was similar to the report obtained in Iran; inpatients and outpatients were 53.0% and 41.0% respectively [27], while different from that of Bosnia-Herzegovina with ESBL prevalence in inpatients of 12.5% [28]. High level ESBL producing isolates were originally confined to inpatients the trajectory of paradigm shift is very high in outpatients and community. This shows the extent of spread and level of resistance in the region. The isolates from outpatient in this case can be regarded as community isolates because the patients were not admitted they only visited these hospitals for treatment which in most cases were brief period spent in the facility. The very high prevalence of ESBL in these hospitals particularly the level in outpatients is a worrisome development as several other reports have indicated overlap between community and nosocomial ESBL.
blaCTX-M variant were the most common observed in this study followed by blaVEB, blaOXA, blaSHV and blaPER. This is in line with other studies showing SHV and TEM families were gradually reducing in prevalence and being supplanted by the CTX-M family [19,29]. The CTX-M family has become widespread in both hospital and community settings virtually in every continents of the world [8,26]. blaCTX-M-15 was the dominant ESBL found among the blaCTX-M variants in accordance with previous reports all over the world [8,26,30]. blaCTX-M-15 has frequently been seen to co-exist with other ESBL genes in the isolates. blaVEB has been reported from most parts of the world since its first description in Vietnam [31] including south-western Nigeria [8,32]. To our knowledge, this was the first report of blaVEB and blaPER ESBLs from northern Nigeria. ESBLs production was found across varying clinical specimens and hospitals.
There was a high level of multidrug resistance to majority of the antibiotics including third-generation cephalosporins, ampicillin, beta-lactamase/inhibitor (amoxicillin-clavulanic acid), fluoroquinolones and aminoglycosides among the isolates. These findings have significant implications in the use of third-generation cephalosporins including fluroquinolones for the management of patients with infections caused by E. coli. In this panel only carbapenems retained good efficacy as a therapeutic option. The low rate of resistance to carbapenems was mirrored by a lack of detection of carbapenemase genes, the most likely scenario may be that carbapenem resistance is being caused by blaCTX-M-15 in conjunction with other mechanisms such as efflux and or porin loss. The absence of carbapenemase genes is very surprising compared to high resistance rates reported from different studies from Southern Nigeria [8,19]. Carbapenem resistance has been reported to be mediated by production of ESBL (largely CTX-M-15) accompanied by porin loss and or efflux activity [33]. The low resistance rate may not be unconnected with the high cost of carbapenem antibiotics which is directly related to the poverty and literacy level in the North of Nigeria in terms of affordability and awareness. Indiscriminate and empirical use of beta-lactam drugs in particular carbapenems should be avoided to guide against the development of carbapenem-resistant strains of E. coli leaving no viable therapeutic options in the future.
Limitation(s)
It was self-sponsored hence it was not possible to investigate further, for instance, confirm mechanisms of few isolates with carbapenem resistance with no carbapenemase genes.
Conclusion(s)
There is a significant challenge in the use of third-generation cephalosporins including other antibiotics except carbapenem in the management of patients with infections caused by E. coli in Nigeria, in the North of the country. This was threatened by widespread dissemination of CTX-M enzymes.
There is need for an effective antibiotic stewardship programme and regular antibiotic resistance surveillance studies enhanced by real time routine detection of ESBLs with a view to stemming the tide of antibiotic resistance and maintain low level resistance especially to carbapenems which remain critically important in this setting.
Numbers in parentheses are percentages
Numbers in parentheses are percentages