In areas with high disease burden, the data on aetiology of pneumonia remains sparse [7]. The identification of the definite aetiology of CAP in blood or sputum samples is challenging despite the progress in diagnostic techniques and sophisticated molecular methods. The expanding rate of emergence of new and multi-drug resistant pathogens globally is a major concern for antibiotic management for CAP [8]. Although the incidence of CAP in India is reported to be 4 million cases annually with mortality rates up to 25% in patients admitted to the intensive care unit, there are very few reports describing the microbial aetiology of CAP [5,9-11]. There is also considerable variation in the microbial patterns, depending on the geographic location, and the use of different types of microbiologic investigations [5,8,11,12].
Knowledge of common microbial patterns in CAP is crucial for making initial therapeutic decisions for empiric antimicrobial treatment. This study was aimed at determining the bacterial aetiology of CAP in patients admitted to a tertiary care hospital in the rural area of Mangalore as many other previous studies have focused on determining the bacterial aetiology of CAP in the urban areas.
Materials and Methods
This was a descriptive study carried out among adult patients, 18 years of age or older, admitted to 1200-bedded tertiary care teaching hospital in the rural area of Mangalore, Karnataka, India with a working diagnosis of CAP. This study was conducted from a period of August 2012 to August 2014 and a total of 220 subjects were enrolled. Subjects with a diagnosis of CAP based on lung consolidation, pleural effusion and infiltrates on chest radiography at the time of admission and ≥2 additional symptoms (temperature >38.3°C, a new cough, chest pain, or new onset of dyspnea) were included in the study [13-15]. Subjects who have been hospitalised within the last 30 days, immunocompromised patients (e.g., HIV/AIDS, organ transplants, cancer patients or recipients of corticosteroids, anti-neoplastic therapy, or other immunosuppressive agents) and subjects with radiographic evidence of pulmonary tuberculosis were excluded [13,15]. All the participants gave written informed consent to participate, and the study protocol was approved by the Central Ethical Committee of the Institution (NU/CEC/Ph.D-46/2012).
Demographic data were collected from patient records which included age, sex, education, occupation, and income. Chest X-ray showing features of pneumonia (lung consolidation, pleural effusion, infiltrates) were noted. Specimens for microbiologic diagnosis included blood and sputum cultures in all enrolled patients. The samples were obtained in a sterile container, and the microscopic evaluation was done by Gram staining. Sputum validity was assessed using the Geckler criteria and samples containing over 10 Squamous Epithelial Cells (SEC) per Low Power Microscope Field (LPMF) were rejected [16]. Tracheal aspirate, pleural fluid, and BAL specimens were collected from the subjects who underwent mechanical ventilation. Samples accepted after microscopic evaluation were inoculated directly onto Sheep blood agar, MacConkey agar and Chocolate agar plates within 30 minutes of sample collection [Table/Fig-1,2] [16]. Culture positive isolates from the samples were initially identified by Gram stain from the colony, colony morphology, and oxidase reaction. They were further subjected to biochemical identification [17].
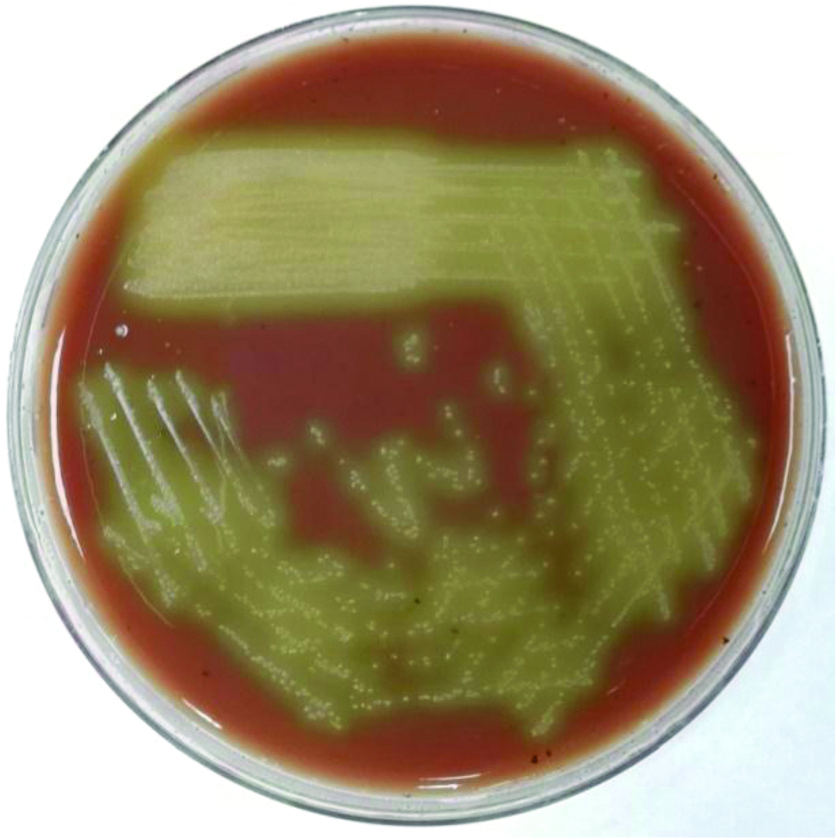
Streptococcus pneumoniae cultured on chocolate agar plate.
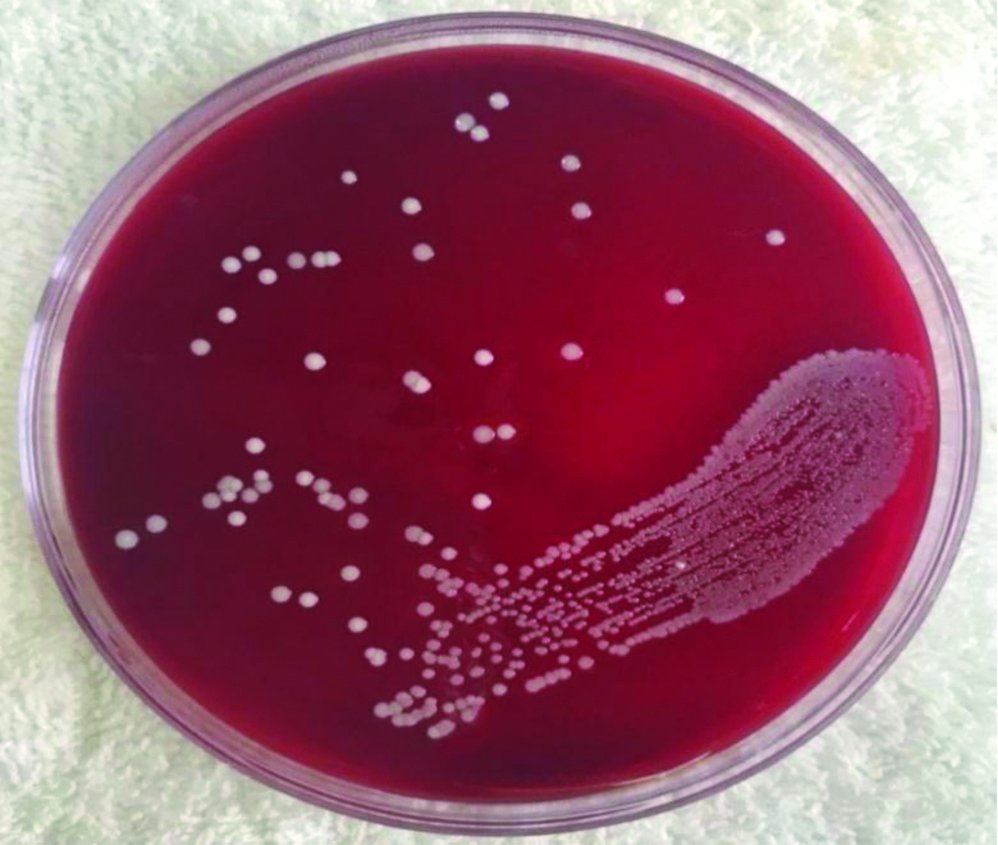
Staphylococcus aureus cultured on sheep blood agar plate.
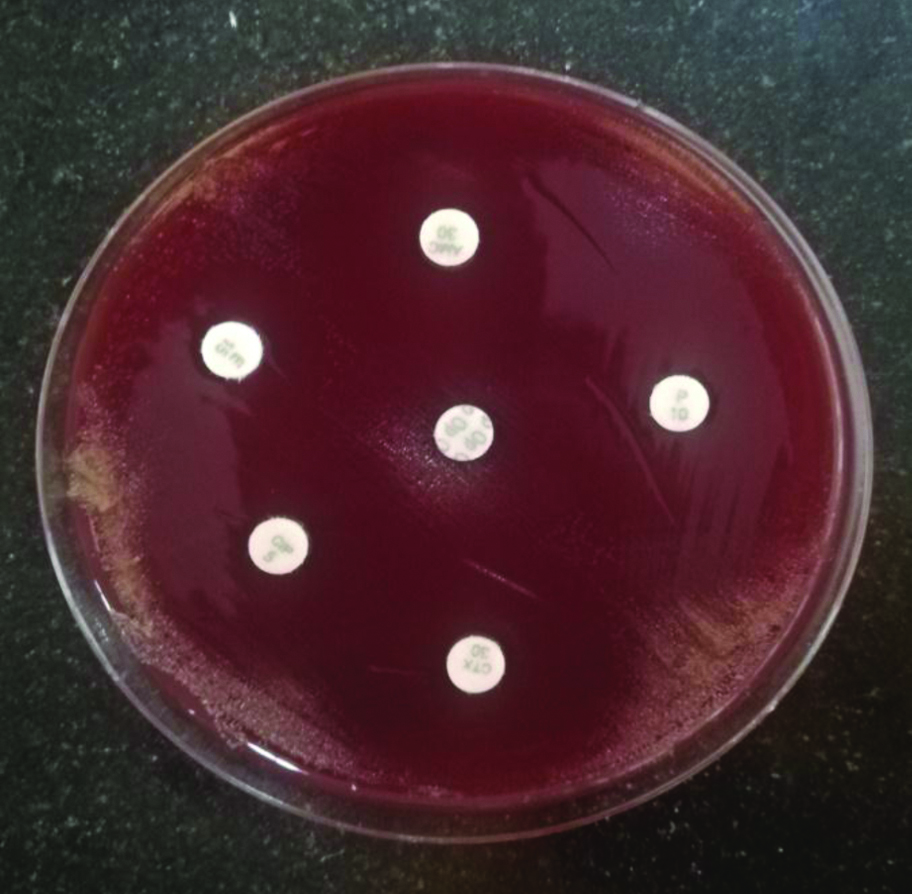
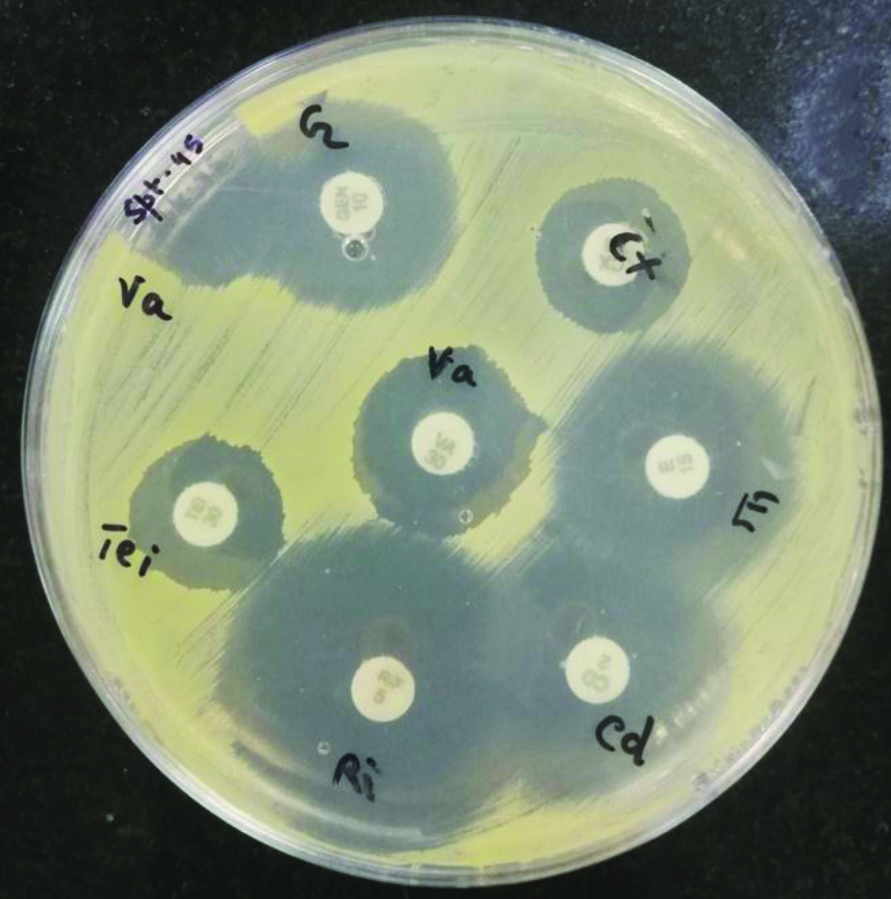
Antibiotic susceptibility testing was done for S.pneumoniae and S.aureus using disk diffusion as described by the Kirby-Bauer method. A single colony from the culture positive isolates of S.pneumoniae and S.aureus were lawn cultured on Sheep blood agar and Mueller-Hinton agar (Himedia Laboratories, India) plates respectively [Table/Fig-3,4]. The plates were incubated at 37°C for 18-24 hours with 5% CO2 for S. pneumoniae. The zone of inhibition was measured, recorded and interpreted as sensitive and resistant according to the CLSI guidelines [18].
Antimicrobial susceptibility testing of Streptococcus pneumoniae by disk diffusion method on sheep blood agar plate.
Antimicrobial susceptibility testing of Staphylococcus aureus by disk diffusion method on Mueller-Hinton agar plate.
LytA (N-acetylmuramoyl-L-alanine-amidase) gene (229 bp), is the major autolytic enzyme of Streptococcus pneumoniae and is responsible for autolysis with great clinical importance. The 16s rRNA gene (538 bp) shows a high level of genetic diversity in H.influenzae. Both the genes were detected from the clinical samples by PCR for further confirmation. The bacterial DNA was extracted using the QIAamp DNA mini kit (QIAGEN-Sample and Assay Technologies). The primers used were as follows:
lytA: 5’-CGGACTACCGCCTTTATATCG-3’ and 5’-GTTTCAATCGTCAAGCCGTT-3’;
16s rRNA: 5’-TCCTAAGAAGAGCTCAGAGAT-3’ and 5’-TGATCCAACCGCAGGTTCC-3’ [19].
Statistical Analysis
Study data was summarised by frequency and percentage. The data were analysed using SPSS (Statistical Package for Social Sciences) version 16.0 software.
Results
Demographic Characteristics
A total of 220 subjects with CAP were enrolled in the study. Out of 220 subjects, bacterial aetiology was determined in 66 (30%) of the CAP cases. Among the 220 CAP cases, 122 (55.4%) were males, and 98 (44.5%) were females [Table/Fig-5]. It was observed that 130 (59%) of the subjects belonged to the age group 55 years and above [Table/Fig-5].
Distribution of the subjects (N=220) according to gender and age.
| Distribution of the subjects according to gender (N=220) | Distribution of the subjects according to age (N=220) |
|---|
| Male (%) | Female (%) | Age group (years) | Culture positive CAP n=66 (%) | Culture negative CAP n=154 (%) |
|---|
| 122 (55.4%) | 98 (44.5%) | 18-25 | 5 (8) | 9 (6) |
| 26-35 | 2 (3) | 5 (3) |
| 36-45 | 6 (9) | 11 (7) |
| 46-55 | 17 (26) | 35 (23) |
| 55 above | 36 (55) | 94 (61) |
Bacterial Aetiology of CAP
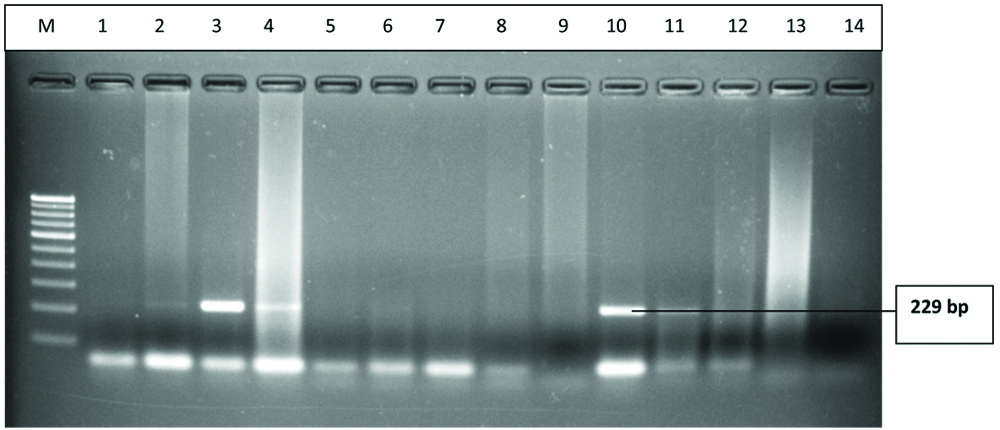
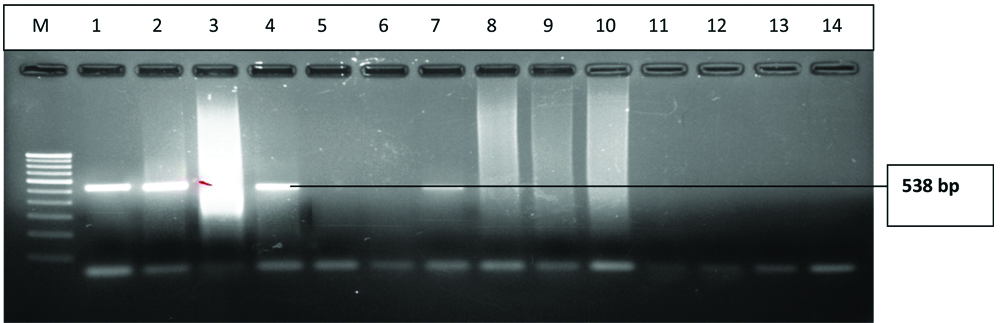
The aetiology of culture positive CAP cases were isolated from various samples received which include: sputum (77%), pleural fluid (18%), tracheal aspirates (3%) and BAL (2%). Of the 66 culture positive cases of CAP, the most commonly isolated pathogens were S. pneumonia (44%) followed by Staphylococcus aureus (32%), H. influenzae (15%), Klebsiella pneumoniae (9%). The lytA gene (229 bp) of S. pneumoniae and the 16s rRNA (538 bp) of H. influenzae was detected by PCR technique. The lanes 3,4,10 and 11 were positive for lytA gene (229 bp) of S. pneumoniae, and the lanes 1,2,4 and 7 was positive for 16s rRNA (538 bp) of H. influenzae respectively [Table/Fig-6,7].
Gel-electrophoresis of PCR amplification of lytA gene: A 229bp fragment specific for lytA gene was amplified: Lane M: 100 bp DNA ladder; Lanes 3,4,10,11: Positive for lytA gene of S.pneumoniae.
Gel-electrophoresis of PCR amplification of 16s rRNA gene: A 538 bp fragment specific for 16SrRNA gene was amplified: Lane M: 100 bp DNA ladder; Lanes 1,2,4,7: Positive for 16s rRNA gene of H.influenze.
Antimicrobial Susceptibility
A 24 (82.75%) of the S. pneumoniae isolates were resistant to penicillin; 6 (21%), 11 (38%), 12 (41%) and 3 (10%) of the S. pneumoniae isolates showed resistance to erythromycin, amoxicillin-clavulanate, ciprofloxacin and cefotaxime respectively [Table/Fig-8].
Antibiotic susceptibility pattern of S.pneumoniae (n=29).
| Name of the antibiotic | Susceptible | Intermediate | Resistant |
|---|
| Amoxicillin/clavulanic acid | 18 | 0 | 11 |
| Cefotaxime | 18 | 8 | 3 |
| Ciprofloxacin | 10 | 7 | 12 |
| Erythromycin | 20 | 3 | 6 |
| Penicillin | 5 | 0 | 24 |
Out of 21, S.aureus isolates, 10 (48%) of them were found to be MRSA. Resistance to clindamycin, erythromycin, rifampicin, and gentamycin was seen among 4 (19%), 8 (38%), 1 (5%) and 8 (38%) of the S.aureus isolates respectively; all S.aureus isolates showed 100% susceptibility to vancomycin and teicoplanin [Table/Fig-9].
Antibiotic susceptibility pattern of S.aureus (n=21).
| Name of the antibiotic | Susceptible | Intermediate | Resistant |
|---|
| Cefoxitin | 11 | 0 | 10 |
| Gentamycin | 13 | 0 | 8 |
| Vancomycin | 21 | 0 | 0 |
| Teicoplanin | 19 | 2 | 0 |
| Rifampicin | 19 | 1 | 1 |
| Erythromycin | 13 | 0 | 8 |
| Clindamycin | 17 | 0 | 4 |
Discussion
This study reported the bacteriological profile of the isolates from adult patients with CAP admitted to a tertiary care hospital in Mangalore, Karnataka, India. A 59% of the subjects with CAP belonged to the age group 55 years and above suggesting that pneumonia is more common in aged individuals than in young adults in the present study setting. This finding is consistent with other studies of CAP in India which showed that the mean age of patients with CAP ranged from 54 to 58 years [9,12]. Studies conducted in high-income countries demonstrate a high incidence of CAP among older adults with substantial morbidity and mortality [20].
In a multicentric, well-conducted prospective study of adult CAP patients needing hospitalisation in the US, a pathogen was detected in only 38% of all patients with CAP [2]. The culture positivity rate of CAP cases in the present study population was only 30%. This may be due to prior antibiotic administration and other aetiological causes of CAP including atypical bacterial and viral pathogens. In contrast, a study conducted in Northern India using standard and molecular diagnostic techniques yielded a higher frequency (72%) of at least one pathogen [5]. In this study, 77% of the culture positive cases were from sputum samples. This result is consistent with a study performed in Ghana wherein 84.9% of the microbes causing CAP were isolated from sputum samples [21].
This study showed that S.pneumoniae was the most common aetiological agent of CAP. In another study from a tertiary care hospital in Mangalore region in Southern India, S.pneumoniae (31%) was reported to be the most predominant bacterial aetiology of CAP [10]. A similar study to assess the microbial aetiology in hospitalised North Indian adults with CAP showed S. pneumoniae (30.5%) as the most common pathogen isolated [5].
In India, there is limited data regarding antimicrobial susceptibility of S. pneumoniae isolated from sterile body fluids (blood, pleural fluid, etc.,) in patients with the invasive pneumococcal disease. A study done in Northern India has shown 50% of S.pneumoniae isolates were resistant to penicillin [22]. 52.6% of S.pneumoniae, resistance to penicillin was demonstrated in a collaborative study done in eight Asian countries including India [23]. A cross-sectional study from Southern India demonstrated emergence of multidrug-resistant strains of S. pneumoniae, isolated from respiratory specimens; in this study, 4% of S.pneumoniae isolated showed total resistance to penicillin and found to be multi-drug resistant strains, whereas, 10% showed intermediate resistance [24]. In the present study, penicillin resistance was demonstrated by 24 (82.75%) of the S. pneumoniae isolates, which is consistent with the findings reported by Chawla K et al., [24]. Another surveillance study evaluating invasive pneumococcal disease among children aged less than five years showed an overall low (8%) penicillin non-susceptibility compared to other studies; erythromycin resistance was 37%, Co-trimoxazole resistance occurred in 66% and 9% of S.pneumoniae isolates were multidrug resistant [25]. Thus, S.pneumoniae resistance profile varies significantly from region to region in India indicating a high level of genetic variation [26,27].
In the present study, MRSA was demonstrated among 10 (48%) of the total CAP cases. None of the isolates showed resistance to vancomycin. This finding is consistent with another report from India [5]. An observational cohort study was done by the Global Initiative for Methicillin-Resistant Staphylococcus aureus Pneumonia (GLIMP) group reported a lower point prevalence of MRSA pneumonia in India (1.4%) compared to the global incidence (3%) [28]. Studies in the US have reported an MRSA prevalence of 2.4% in CAP [29]. However, community acquired MRSA infections can manifest as severe necrotising pneumonia with high morbidity and mortality, especially following influenza infection [30,31]. In a study reported by Eshwara VK et al., bacteraemic pneumonia due to community acquired S.aureus infections were characterised by a severe disease with high case fatality rate, similar to other studies from high-income countries [32,33]. Similar to present study results, other reports from India have also shown increased rates of resistance of S. aureus isolates to ciprofloxacin and erythromycin. Therefore, the choice of empiric antibiotic therapy for suspected invasive S.aureus infections remains a major challenge in this study setting [33].
Molecular methods like PCR emphasises the effective identification of respiratory bacterial pathogens. Screening for the typical pathogens of CAP such as S.pneumoniae and H.influenzae were confirmed by PCR techniques in the present study. LytA (N-acetylmuramoyl-L-alanine-amidase), is the major autolytic enzyme of Pneumococcus being responsible for the deoxycholate and penicillin induced cell lysis in the stationary phase with great clinical importance [34]. The 16s rRNA gene diversity showed a higher level of diversity in H.influenzae ranging from 0.6% to 2.73% as reported by Sacchi CT et al., [35]. The detection of both lytA gene (229 bp) and the 16s rRNA (538 bp) of S.pneumoniae and H.influenzae respectively in this study showed that PCR is a highly sensitive and specific technique in identifying the bacterial aetiology of lower respiratory tract infections.
Future studies should evaluate the influence of age, co-morbidity, and disease severity on the microbial pattern of CAP. Due to the emergence of resistant S.pneumoniae and S.aureus- two key pathogens causing CAP, establishment of antimicrobial stewardship, infection control program and ongoing antimicrobial resistance surveillance is a key to guide clinicians and policy makers about optimal antibiotic management of CAP in the present study setting [36].
Limitation(s)
This study had several limitations. First, the occupation and income of the enrolled CAP subjects were not reported and also the impact of other factors such as co-morbidity, severity on the microbial patterns associated with CAP were not assessed due to unavailability of the complete clinical profile and demographic data among all patients. Second, even though a blood culture was obtained among all enrolled subjects, the blood culture results for all pathogens isolated were not mentioned due to a very low yield of bacterial CAP pathogens and higher frequency of possible contaminants (e.g., coagulase-negative Staphylococcus species). Third, diagnostic testing for atypical bacterial pathogens (e.g., Legionella and Mycoplasma) and viral pathogens were not carried out due to lack of funding. Fourth, the antimicrobial susceptibility testing was done only for S.pneumoniae and S.aureus isolates with a limited number of antibiotics and the antibiotic susceptibility testing was not carried out for the Gram negative isolates as the study focused on the resistance profile of Gram positive isolates. Finally, this study was performed in the in-patient setting at a single, tertiary care hospital in the rural area of Mangalore and results may not be generalised to other regions in India.
Conclusion(s)
CAP commonly affects the aged population than in young adults. Culture techniques help in identifying 1/3rd of the aetiology of CAP. PCR may prove to be a rapid and sensitive technique in the detection of CAP pathogens directly from the clinical samples. The prevalence of S.pneumoniae and S.aureus was higher compared to Gram negative bacterial aetiology among CAP patients in the rural area of coastal city in Southern India.