Infections of skin and soft tissue can appear from the incursion of organisms from the external environment through skin breakage or from organisms that get to the skin via blood as a part of systemic illness [1]. These can result into pyogenic infection characterised by severe local inflammation, usually with pus formation caused by one of pyogenic bacteria, which can produce accumulation of dead leucocytes and infectious agent [2].
Pyogenic infection can be a chief complication of surgery and trauma; these are the main cause of morbidity and mortality. Factors that are contributing to these infections are pre-existing illness, length of operation, length of stay in hospital, wound class and wound contamination [3]. These infections can be exogenous or endogenous or it may be monomicrobial or polymicrobial in nature [4,5].
Increasing antibiotic resistance among the various isolates of skin and soft tissue infection is alarming [2,3,5,6]. Hence, there is a need for identification of causative organism and their antimicrobial susceptibility pattern. This is required both at hospital and community level on regular basis to guide the clinicians regarding judicious and rational usage of antibiotics. Studies across the globe, suggest that aerobic bacteria cause pyogenic infections [2,3,6]. Hence, the present study was undertaken to establish the aerobic bacterial profile and antimicrobial susceptibility pattern of various isolates obtained from wounds, abscesses, tissues and tissue aspirates in a tertiary care teaching hospital.
Materials and Methods
This retrospective study was conducted in the Department of Microbiology over a period of one year from January to December 2017. Records for a total of 880 samples received for aerobic culture from various wards, ICU and OPD, were included in the study. Anaerobic bacteria were excluded from the study.
The samples included pus, pus swab, wound swab, tissue and tissue aspirates. As per the hospital protocol, the samples were simultaneously subjected to Gram stain and aerobic culture. They were inoculated on Blood agar and MacConkey agar followed by overnight incubation at 37°C. Identification of the organisms were done by standard microbiological techniques [7].
Antimicrobial susceptibility testing was done by modified Kirby Bauer’s disc diffusion method and interpretation was done by CLSI guidelines 2016 [8]. Depending on the isolate the antibiotic disc were selected from the following, Amoxycillin-clavulanic acid (20/10 μg), Ceftriaxone (30 μg), Ceftazidime (30 μg), Gentamicin (10 μg), Amikacin (30 μg), Ciprofloxacin (5 μg), Cotrimoxazole (25 μg), Piperacillin-Tazobactum (100/10 μg), Meropenem (10 μg), Ampicillin (10 μg), Erythromycin (15 μg), Clindamycin (2 μg), High Level Gentamicin (HLG) (120 μg), Vancomycin (30 μg) and Linezolid (30 μg).
Statistical Analysis
The socio-demographic data (gender, wards, pus culture results and their antimicrobial susceptibility pattern) were taken manually from laboratory register and analysed in counts and percentages using MS Excel 2010 version.
Results
Out of 880 samples, majority 470 (53.41%) were received from the Department of Surgery and the least number of samples were received from ICU [Table/Fig-1]. Out of 880 samples, 541 (61.48%) samples were received from males and 339 (38.52%) were received from females. Growth was obtained in 574 (65.23%) samples in which 38 (6.62%) samples showed polymicrobial growth. The total number of isolates were 612 (69.54%) in which 375 (61.27%) were gram negative bacteria [Table/Fig-2] and 233 (38.07%) were gram positive bacteria [Table/Fig-3] whereas 04 (0.65%) isolates were non-albicans Candida species. Among the gram negative bacteria, most common isolate was Pseudomonas aeruginosa 110 (29.33%) followed by Escherichia coli 99 (26.4%), whereas Methicillin Sensitive Staphylococcus Aureus (MSSA) 75 (32.19%) predominated among the gram positive bacteria followed by MRSA 59 (25.32%). However, overall the most common bacterial organism was Staphylococcus aureus 134 (22.04%) followed by Pseudomonas aeruginosa 110 (18.09%) and other bacterial pathogens [Table/Fig-4]. The antibiotic sensitivity of Pseudomonas aeruginosa displayed that majority of them were sensitive to Meropenem 85 (77.27%) followed by Piperacillin-Tazobactum 72 (65.45%) [Table/Fig-5]. All gram positive cocci were sensitive to Vancomycin and Linezolid [Table/Fig-6].
Speciality wise distribution of samples (n=880).
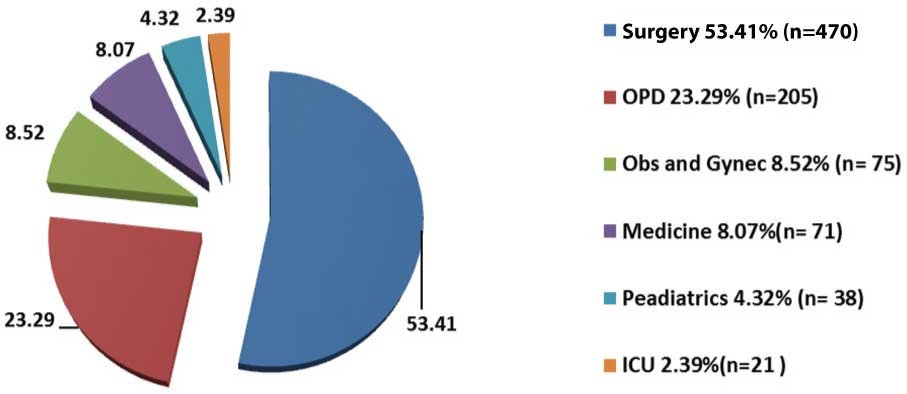
Percentage distribution of gram negative organisms (n=375).
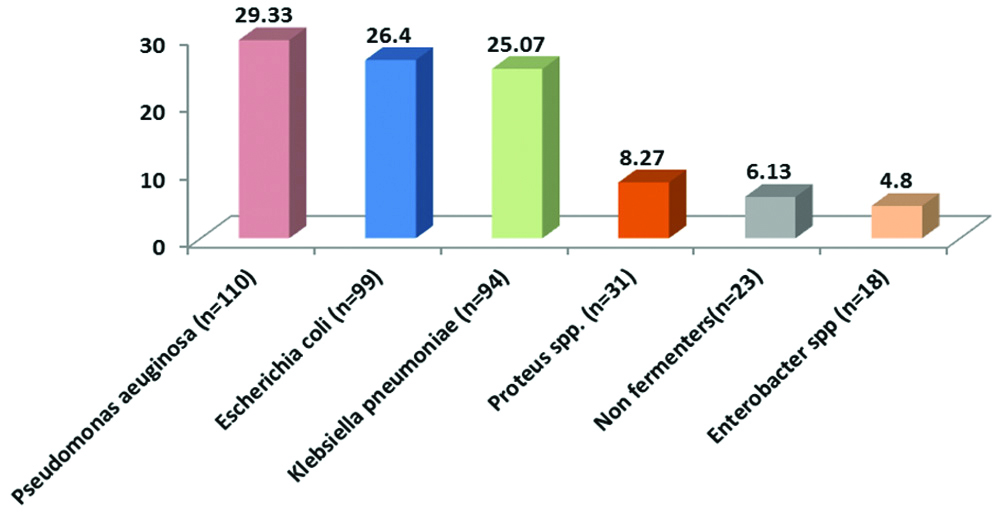
Percentage distribution of gram positive organisms (n=233).
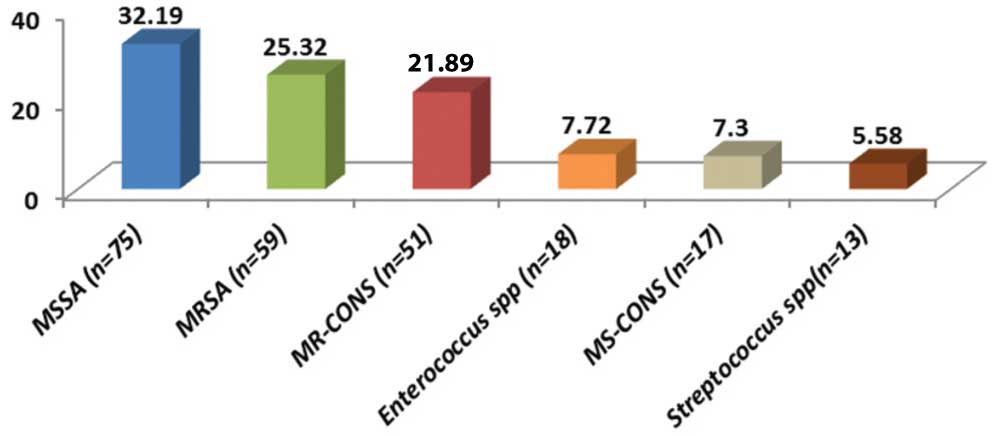
Percentage distribution of bacterial isolates (n=608).
Antibiotic susceptibility pattern of gram negative organisms.
| Antibiotics (μg /disc) | Pseudomonas aeruginosa (n=110) | Escherichia coli (n=99) | Klebsiella pneumoniae (n=94) | Proteus spp. (n=31) | Non fermenter (n=23) | Enterobacter spp (n=18) |
|---|
| Amoxycillin-clavulanic acid (20/10 μg) | 29.09% (n=32) | 11.11% (n=11) | 8.51% (n=8) | 96.77% (n=30) | 8.69% (n=2) | 22.22% (n=4) |
| Ceftriaxone (30 μg) | 27.27% (n=30) | 21.21% (n=21) | 25.53% (n=24) | 61.29% (n=19) | 30.43% (n=7) | 38.89% (n=7) |
| Ceftazidime (30 μg) | 53.64% (n=59) | -- | -- | -- | -- | -- |
| Gentamicin (10 μg) | 58.2% (n=64) | 57.57% (n=57) | 37.23% (n=35) | 54.84% (n=17) | 34.78% (n=8) | 55.55% (n=10) |
| Amikacin (30 μg) | 63.64% (n=70) | 53.53% (n=53) | 31.91% (n=30) | 41.94% (n=13) | 21.74% (n=5) | 33.33% (n=6) |
| Ciprofloxacin (5 μg) | 61.82% (n=68) | 42.42% (n=42) | 31.91% (n=30) | 48.39% (n=15) | 30.43% (n=7) | 38.89% (n=7) |
| Cotrimoxazole (25 μg) | 18.18% (n=20) | 27.27% (n=27) | 34.04% (n=32) | 22.58% (n=7) | 52.17% (n=12) | 61.11% (n=11) |
| Piperacillin-Tazobactum (100/10 μg) | 65.45% (n=72) | 54.54% (n=54) | 38.30% (n=36) | 67.74% (n=21) | 17.39% (n=4) | 38.89% (n=7) |
| Meropenem (10 μg) | 77.27% (n=85) | 61.61% (n=61) | 48.94% (n=46) | 74.19% (n=23) | 43.48% (n=10) | 50% (n=9) |
Antibiotic susceptibility pattern of gram positive organisms.
| Antibiotics (μg /disc) | MSSA (n=75) | MRSA (n=59) | MR-CONS (n=51) | Enterococcus spp (n=18) | MS-CONS (n=17) | Streptococcus spp (n=13) |
|---|
| Ampicillin (10 μg) | 45.33% (n=34) | 0% | 0% | 44.44% (n=8) | 47.06% (n=8) | 76.92% (n=10) |
| Cotrimoxazole (25 μg) | 50.67% (n=38) | 64.41% (n=38) | 52.94% (n=27) | 50% (n=9) | 64.70% (n=11) | -- |
| Ceftriaxone (30 μg) | 69.33% (n=52) | 27.12% (n=16) | 39.22% (n=20) | 44.44% (n=8) | 70.59% (n=12) | 46.15 (n=6) |
| Erythromycin (15 μg) | 56% (n=42) | 27.12% (n=16) | 27.45 (n=14) | 33.33% (n=6) | 47.06% (n=8) | 69.23% (n=9) |
| Clindamycin (2 μg) | 72% (n=54) | 44.07% (n=26) | 52.94% (n=27) | 55.55% (n=10) | 76.47% (n=13) | 53.85% (n=7) |
| Gentamicin (10 μg) | 76% (n=57) | 50.85% (n=30) | 49.02% (n=25) | 27.78% (n=5) | 70.59% (n=12) | 38.46% (n=5) |
| Ciprofloxacin (5 μg) | 65.33% (n=49) | 22.03% (n=13) | 35.29% (n=18) | 33.33% (n=6) | 64.70% (n=11) | 38.46% (n=5) |
| HLG (High Level Gentamicin) (120 μg) | -- | -- | -- | 22.20% (n=4) | -- | -- |
| Vancomycin (30 μg) | 100% (n=75) | 100% (n=59) | 100% (n=51) | 100% (n=18) | 100% (n=17) | 100% (n=13) |
| Linezolid (30 ug) | 100% (n=75) | 100% (n=59) | 100% (n=51) | 100% (n=18) | 100% (n=17) | 100% (n=13) |
Discussion
In the present study, majority of the samples were received from surgery department 470 (53.41%). Higher number of samples from surgery department has been observed in many studies conducted on pus samples such as Siddiqui F et al., {54 (72%)}, Roopa C and Deepali V {198(67.5%)}, Karia JB et al., {154 (59.45%)}, Biradar A et al., {93(55.2%)} and Duggal S et al., {36 (32.43%)} [6,9-12]. The reason for this could be because pus and wound discharge cases mostly present to surgery department. Samples received from males {541 (61.48%)} were greater as compared to females {339 (38.52%)}, since the category of patients received in our hospital are mostly labourers by occupation and males are usually involved in such activities, they are more prone to develop wounds and abscesses. This correlated with studies done by Siddiqui F et al., 18 (72%) and Roopa C and Deepali V 184 (63%) which also shows male preponderance [9, 10]. During the study period, out of the total 880 samples 574 (65.23%) were positive for growth. This isolation rate correlates with studies done on pus culture in India by Roopa C and Deepali V, Biradar A et al., and Trojan R et al., showing isolation rates of 60.40% (177/293), 66.01% (169/256) and 60.1% (86/143) respectively [10,11,13]. A study done by Muluye D et al., in Africa also showed a similar isolation rate of 70.2% (441/628) [14]. Differences in isolation rates may be due to differences in the time of receiving and processing of samples in various laboratories. Out of the total 574 samples, 38 (6.62%) showed polymicrobial growth, which is showing similarity with the study done by Roopa C and Deepali V 11 (6.21%) and Biradar A et al., 12 (7.1%) [10,11].
The total number of isolates was 612 (69.54%) in which 375 (61.27%) were gram negative bacteria, 233 (38.07%) were gram positive bacteria whereas 04 (0.65%) isolates were non-albicans Candida species. Predominance of gram negative bacteria correlates well with study by Roopa C and Deepali V, who had 112 isolates of gram negative bacteria (60%) [10] and Hanumanthappa P et al., with 157 isolates (54.32%) [15]. Minimal isolation rate of fungal organisms has also been observed in studies done by Karia JB et al., 2 (1.36%) [6], Roopa C and Deepali V, 1 (0.53%) [10] and Hanumanthappa P et al., 4 (0.8%) [15] which was similar to the present study.
In the present study, overall the most common bacterial organism was Staphylococcus aureus 134 (22.04%). Similar finding has been observed in Indian studies by Subha M and Srinivasagam M {80(26.32%)} [16], Sudhaharan S et al., {443(29.05%)} [17] and Verma P {46 (40%)} [18] conducted in Tamil Nadu, Telangana and Chhattisgarh respectively. Also, studies done outside India by Khanam RA et al., [19] in Bangladesh {53 (25%)} and Rai S et al., [20] in Nepal {160 (60.60%)} showed Staphylococcus aureus to be the predominant pathogen. Among the gram negative isolates, the most common pathogens was Pseudomonas aeruginosa 110 (29.33%) followed by E. coli 99 (26.4%). Similarly, P. aeruginosa was the most common gram negative isolate in studies done by Duggal S et al., {33 (29.73%) [12] and Rai S et al., {45 (44%)} [20]. However, this is in contrast with the studies done by Khanam RA et al., {35 (16.5%)} [19], Subha M et al., {42 (13.82%)} [16] and Sudhaharan S et al., {403 (38.6%)} [17] where Escherichia coli was found to be the predominant gram negative organism. The differences in the distribution of bacterial isolates may be attributed to the differences in study design, geographical location and climatic conditions.
Out of the total 134 isolates of S. aureus, 75 (55.97%) were MSSA and 59 (44.03%) were MRSA. This correlates with that of Sudhaharan S et al., who isolated 251 (51.9%) MSSA and 192 (39.7%) MRSA strains [17]. All S. aureus isolates were 100% sensitive to Vancomycin and Linezolid which is in accordance with the study done by Roopa C and Deepali V [10] and Rameshkannan S et al., [21] showing 100% sensitivity towards Vancomycin and Linezolid. Among all MSSA isolates, 76% (57) were sensitive to Gentamicin, 65.33% (49) were sensitive to Ciprofloxacin and 56% (42) were sensitive to Erythromycin. Similar findings were noted in the study by Subha M and Srinivasagam M in which 75%, 63.75% and 68.75% S. aureus isolates were sensitive to Gentamicin, Ciprofloxacin and Erythromycin respectively [16].
Most gram negative isolates were sensitive to Meropenem followed by Piperacillin-Tazobactum, Ciprofloxacin and Aminoglycosides while they were less sensitive to Amoxycillin-Clavulanic acid, Cephalosporins and Cotrimoxazole. Among the isolates of Pseudomonas aeruginosa (the most common gram negative organism in this study), 77.27% (85) were sensitive to Meropenem, followed by Piperacillin-Tazobactam {65.5% (72)}, Amikacin {63.64% (70)} and Ciprofloxacin {61.82% (68)}. Similar findings were noted in the study by Biradar A et al., where 64.48% (31) isolates of Pseudomonas aeruginosa were sensitive to Meropenem, 60.41% (29) isolates to Piperacillin-Tazobactam, 52.08% (25) were sensitive to Amikacin and 37.5% (18) were sensitive to Ciprofloxacin [11]. However, in the study by Roopa C and Deepali V, all isolates (16) of Pseudomonas aeruginosa were sensitive to Imipenem followed by Piperacillin-Tazobactam (87.5%) and Amikacin (68.75%) [10].
Since, multidrug resistant bacteria are emerging worldwide which cause public health problems and challenges to health care, knowledge of the most common causative agents of infection and their antimicrobial susceptibility pattern is very essential for the judicious administration of empirical therapy before culture results are available [2,6].
Similar studies should be carried out on a larger scale and on regular basis to know the trend of antimicrobial resistance pattern present in the community which can guide surgeons to select the empirical antibiotics judiciously to avoid development of antimicrobial resistance. Proper wound care and its management along with proper education regarding strict use of infection control measures such as hand hygiene practices in such patients can avoid wound infection and its spread through contaminated hands in the environment. This can reduce the burden of resistant organisms both in the hospital and the community.
Limitation(s)
Minimum Inhibitory Concentration (MIC) value of antibiotics could not be determined due to unavailability of E-strips and other resources. Molecular level analysis of the isolates and their typing could not be done.
Conclusion(s)
The most common causative organism obtained from various samples of pus was Staphylococcus aureus. All gram positive bacteria were sensitive to Vancomycin and Linezolid. Hence, Vancomycin and Linezolid were the most promising drugs against gram positive bacteria. The most common gram negative pathogen was Pseudomonas aeruginosa and the majority of gram negative pathogens were sensitive to Meropenem and Piperacillin-Tazobactum. Since antibiotic resistance is a rising concern this study can help clinicians to select and use the antibiotics judiciously, preventing the development of further antibiotic resistance among our hospital isolates.