The use of antibiotics for treatment of diseases in both human and animals started before 80 years and the excessive and uncontrolled use resulted in development of antibiotic resistance [1]. It is the ability of a microbe to survive and multiply in presence of antibiotics which is transmissible through environmental pathways and has become a universal threat to human and animals [2]. Antibiotic resistant genes from MDR bacteria are generally transferred by plasmid conjugation to other bacterial strains [3]. The sea shores are contaminated with xenobiotic components such as industrial effluents, hospital and domestic wastes which contain herbicides, pesticides, heavy metals and antibiotics. Presence of such components in water along with excessive usage of chemicals and antibiotic in sea foods exerts pressure on bacteria for survival and to get mutated and becomes resistant to antibiotics [4]. Antibiotic residues in sea foods are persistent organic pollutant which get introduced into human diet which alter the normal human microbiome to become resistant to antibiotics, making a reservoir for resistant genes [5]. Thus, choice of antibiotics to treat infections becomes limited and challenging making antibacterial chemotherapy, difficult and expensive [6,7].
Sea foods are relatively free of pathogenic microorganisms which are later contaminated by pathogens during handling, post-harvest process and during storage [8,9]. Recently, due to the extensive use of antibiotics, there are reports on contamination of sea food with MDR bacteria having Extended Spectrum Beta-Lactamases (ESBL) that confer resistance to β-lactam antibiotics [10]. It is commonly seen in E.coli, Klebsiella and Proteus sp. however, other Gram negative bacteria such as Aeromonas and Enterobacter were also reported to produce such enzymes [11]. Members of Enterobacteriaceae Vibrionaceae, Pseudomonadaceae and other bacteria such as Roseateles aquatilis, Photobacterium damselae which causes serious food borne illness were isolated from marine fish samples collected from South Jeddah, Saudi Arabia [12]. Bacteria including Klebsiella, Pseudomonas, and Methicillin resistant Staphylococcus and Streptococcus species were also detected from sewage collected from fish market [13]. Fishes sold in market are carrier for pathogenic bacteria and rapid increase in the MDR bacteria in food chain is alarming and is a serious threat to public. There are bacterial pathogens in fishes which can be isolated apparently without any symptoms. The safety in consuming sea food sourced from informal markets is unknown and it may lead to food borne illness. Therefore, there is an urgent need for investigating the occurrence of pathogenic MDR bacteria in the seafood. In the present study, fish samples were collected from local market and the samples were subjected to bacterial analysis and antibiotic sensitivity test to find MDR.
Materials and Methods
Sample Collection
This was a random study conducted at College of Applied Medical Sciences, Shaqra, Kingdom of Saudi Arabia. The study was conducted in October to December 2018 for a period of three months and 10 samples were collected in each month. Fresh fish samples (n=30) were collected randomly from the local fish market at Shaqra, Riyadh province. Small fishes kept in open space in ice were taken as samples whereas, large fishes and fishes stored in freezers were not used for the study. Fresh fish samples were collected in sterile plastic bags from the market and transported in ice to the laboratory under aseptic conditions at 4°C. The samples were processed immediately for further studies. The processing of fish samples were done according to the standard method [14]. The experimental procedure and the scientific content of the research were approved by the Institute Ethical Committee (SU/CAMS/15/12-2014/2).
Isolation of Bacteria
Bacteria were isolated from the samples on two different bacteriological culture media such as MacConkey agar and blood agar. After inoculation, the culture plates were incubated at 37°C for 18-24 hours. Colonies on blood agar were observed for haemolysis pattern. MacConkey agar was used as a selective differential media for differentiating lactose fermenters from non-lactose fermenters. Eighty morphologically different colonies found commonly in all samples were selected for the further studies. The Gram negative bacterial isolates were identified based on colony morphology, Gram’s staining, motility test and biochemical reactions by using Analytical Profile Index (API 20E and API 20NE, Biomerieux, France) [15]. Gram positive isolates were identified using morphological and biochemical characteristics [16].
Antibiotic Sensitivity Test
All 80 isolates of bacteria were subjected to antibiotic sensitivity test using Kirby-Bauer disc diffusion method. Antibiotics chosen for the present study includes amoxicillin (10 mcg), bacitracin (10 mcg), chloramphenicol (30 mcg), ciprofloxacin (5 mcg), erythromycin (15 mcg), gentamycin (10 mcg) and tetracycline (30 mcg). Muller Hinton Agar medium was prepared, sterilised and poured into petri-dishes and stored at 4°C until use. Fresh bacterial cultures were prepared according to 0.5 McFarland’s standard turbidity. The bacterial suspension was swabbed using sterile cotton swab over the entire surface of the medium. After the inoculum was dried, commercially available antibiotic discs were applied with forceps and pressed gently to ensure even contact with medium. Plates were incubated for 18 to 24 hours at 37°C aerobically. After incubation, diameters of complete inhibition zones formed around the disc were measured to the nearest whole millimetre using a standard ruler. The results were analysed and interpreted according to the reference tables provided by Clinical and Laboratory Standards Institute (CLSI), standards, 2016 [17].
Multiple Antibiotic Resistance Indexing
The Multiple Antibiotic Resistance (MAR) indexing for the same bacterial genera isolated from the fish samples were calculated. The index was derived using the formulae a/b*c; where ‘a’ is the aggregate antibiotic resistance score of all isolates belonging to same genera, ‘b’ is the number of antibiotic used in the study and ‘c’ is the total number of isolates from the sample [18]. For example, if 15 (c) E. coli isolates were isolated, the total aggregate antibiotic resistance score is 47 (a) and the total number of antibiotics used was 7 (b), the MAR index would be 47/(7×15) or 0.44.
Statistical Analysis
The obtained research data were analysed by SPSS 23 version and it was summarised by percentage and data presented in mean±standard error.
Results
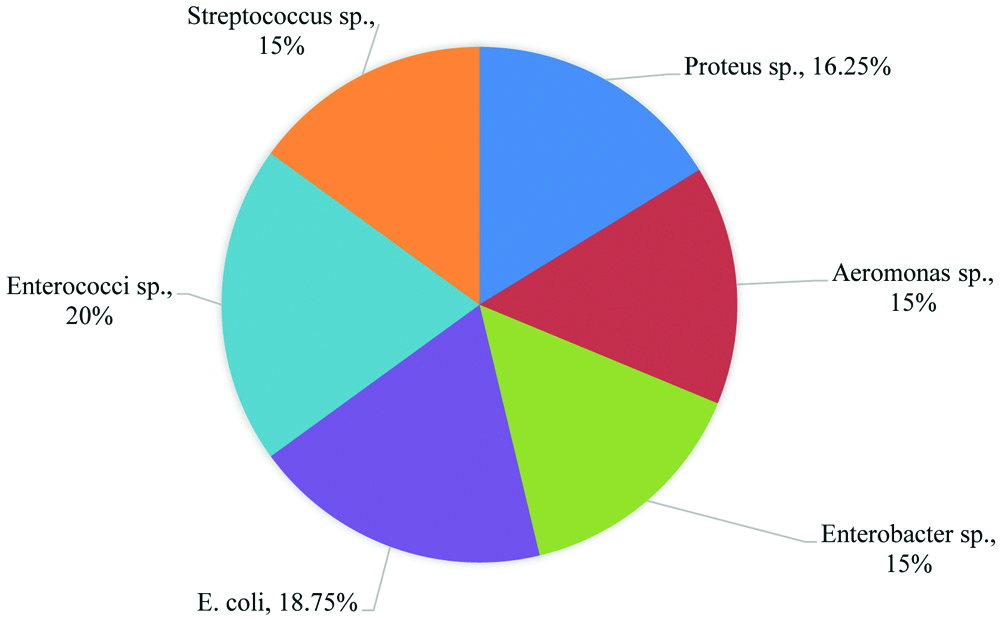
In the present study, randomly collected 30 fish samples were analysed for bacterial contamination. From the culture plates morphologically different colonies were isolated and made as pure culture by repeated streaking into nutrient agar. Proteus species (8), E. coli (10), Aeromonas species (7) and Enterobacter (8) and Enterococcus (8) were isolated from MacConkey agar. Proteus species (5), E. coli (5), Aeromonas species (5) Enterobacter (4) Enterococcus (8) and Streptococcus (12) were isolated from blood agar. Among the 80 isolates, 28 isolates (35%) were Gram positive and 52 isolates (65%) were Gram negative bacteria. Out of 28 Gram positive bacteria, 16 (20%) isolates were able to tolerate 45°C temperature, 6.5% NaCl and 40% bile salt during growth. It showed α-haemolysis and positive reaction for bile esculin test. Based on biochemical characteristics, these isolate were identified as Enterococcus sp. The remaining 12 (15%) isolates were identified as Streptococcus sp. based on morphological and biochemical characteristics. All other Gram negative isolates were identified based on morphological and biochemical tests by API 20 E test at genus level. The isolates include Proteus sp. 13 isolates, Escherichia coli 15 isolates, Enterobacter sp. 12 isolates and Aeromonas sp. 12 isolates [Table/Fig-1].
Number of bacteria isolated from fish samples.
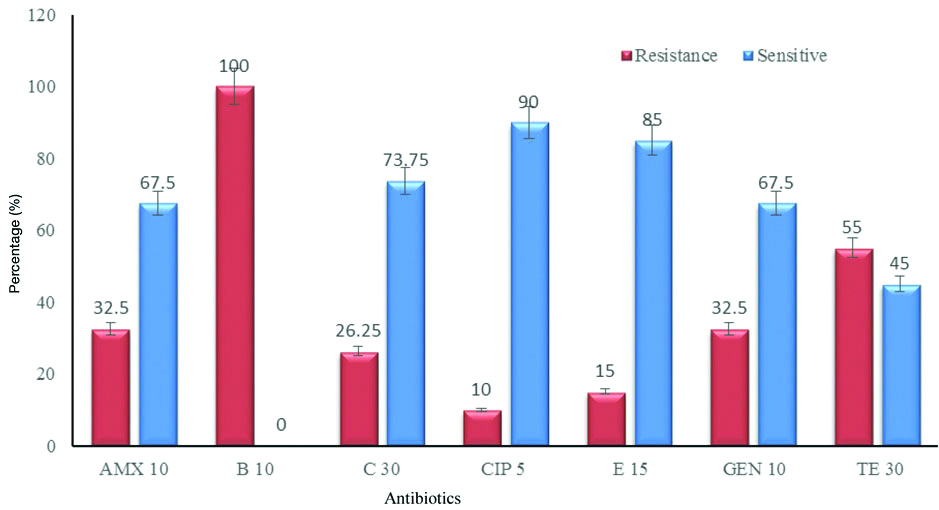
The representative bacterial isolates are characterised by varying differences in the level of resistance [Table/Fig-2]. All Gram negative strains of Proteus species, E. coli, Aeromonas species and Enterobacter species were resistant to bacitracin. Proteus species (7), E. coli (10), Aeromonas species (7) and Enterobacter (5) showed resistance to tetracycline. Only few isolates of these strains were resistant towards amoxicillin, gentamycin and erythromycin. Whereas, only one strain of each Proteus, Aeromonas and Enterobacter were resistant to ciprofloxacin.
Antibiotic resistance shown by the isolates.
| Pathogens | No. of isolates showing resistance (%) |
|---|
| AMX | B | C | CIP | E | GEN | TE |
|---|
| Proteus sp. (n=13) | 38.4 | 100 | 30.7 | 7.6 | 15.3 | 38.4 | 53.8 |
| Escherichia coli (n=15) | 46.6 | 100 | 26.6 | 13.3 | 20.0 | 40.0 | 66.6 |
| Aeromonas sp. (n=12) | 33.3 | 100 | 25.0 | 8.3 | 8.3 | 25.0 | 58.3 |
| Enterobacter sp. (n=12) | 33.3 | 100 | 16.6 | 8.3 | 16.6 | 33.3 | 41.6 |
| Enterococcus sp. (n=16) | 25.0 | 100 | 31.2 | 12.5 | 18.7 | 18.7 | 68.7 |
| Streptococcus sp. (n=12) | 16.6 | 100 | 25.0 | 8.3 | 8.3 | 41.6 | 33.3 |
AMX: Amoxcillin (10 μg); B: Bacitracin (10 μg); C: Chloramphenicol (30 μg); CIP: Ciprofloxacin (5 μg); E: Erythromycin (15 μg); GEN: Gentamycin (10 μg); TE: Tetracycline (30 μg)
The antibiotic resistance of Gram positive bacteria were also studied. All Enterococcus (16) and Streptococcus species (12) showed resistance to bacitracin. Majority of the strains showed intermediate inhibitory or resistance towards gentamycin and tetracycline. Only few strains were resistant to ciprofloxacin, amoxicillin and erythromycin.
Increased resistance pattern of bacteria may be due to the genetic variation or presence of R-plasmid and expression of genes encoding for multi-drug efflux pump which removes wide range of drugs. In the present study, bacteria resistant to four or more antibiotics are considered as MDR strains. All the representative bacterial genera showed resistant pattern against four or more antibiotics [Table/Fig-3]. It is notable that a single strain of bacteria did not show resistance to all the seven antibiotics tested, however, some of the strains showed a maximum intermediate or resistance to five antibiotics used in the study. Escherichia coli (6), Proteus (5), Enterobacter species (4) and Aeromonas (3) were resistant towards four antibiotics used in the study. Gram positive bacteria, Enterococcus (4) and Streptococcus (3) also showed multiple drug resistance. The 80 isolates from fish samples were analysed for multiple antibiotic resistant pattern and 26 (32.5%) of the isolates were resistant to amoxicillin and gentamycin and all the strains were resistant to bacitracin [Table/Fig-4]. More than 50% of the bacterial isolates were resistant to tetracycline.
Percentage of bacterial species showing multidrug resistance.
| Bacterial isolates | No. of antibiotics |
|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|
| Proteus sp. (n=13) | 100 | 53.8 | 38.4 | 38.4 | 30.7 | 15.3 | 7.6 |
| Escherichia coli (n=15) | 100 | 66.6 | 46.6 | 40 | 26.6 | 20 | 13.3 |
| Aeromonas sp. (n=12) | 100 | 58.3 | 33.3 | 25.0 | 25.0 | 8.3 | 8.3 |
| Enterobacter sp. (n=12) | 100 | 41.6 | 33.3 | 33.3 | 16.6 | 16.6 | 8.3 |
| Enterococcus sp. (n=16) | 100 | 68.7 | 31.2 | 25.0 | 18.7 | 18.7 | 12.5 |
| Streptococcus sp. (n=12) | 100 | 41.6 | 33.3 | 25.0 | 16.6 | 8.3 | 8.3 |
Antibiotic resistance pattern shown by the bacteria isolated form the fish samples (n=80).
AMX: Amoxcillin (10 μg); B: Bacitracin (10 μg); C: Chloramphenicol (30 μg); CIP: Ciprofloxacin (5 μg); E: Erythromycin (15 μg); GEN: Gentamycin (10 μg); TE: Tetracycline (30 μg)
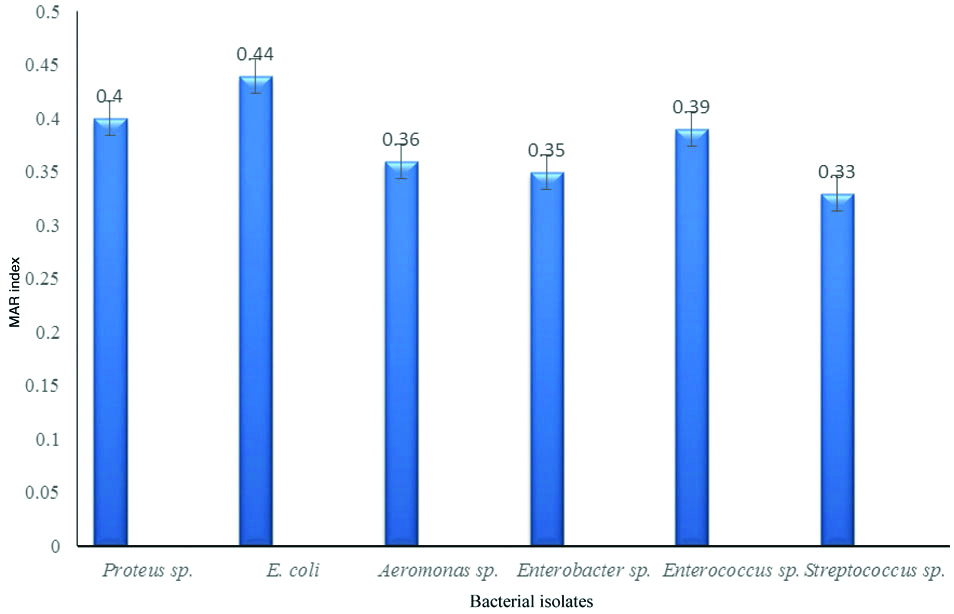
The low and high risk bacterial pathogens are also determined by MAR index which is 0.2 or more. These bacterial isolates are originated from highly contaminated sites and needs careful attention. In the present study, most of the genera showed high MAR index [Table/Fig-5]. E.coli showed the highest MAR index of 0.44 followed by Proteus and Enterococcus species.
MAR index of different bacterial isolates from fish samples.
Discussion
All the samples showed visible growth on agar plates. The occurrence of E. coli in the fish samples is considerably high when compared to other Gram negative bacteria, such as Proteus species followed by Aeromonas and Enterobacter. Fish contaminated with bacteria causing food borne illness has been reported in Saudi Arabia [12]. It is due to unhygienic handling, water or ice used for the storage of fish or contamination of sea water. The presence of coliforms in fish samples demonstrated the level of pollution of their environment because the coliforms are not found commonly in normal fish. The presence of E. coli in fishes may be due to handling or from the faecal contaminated water. Advanced purification techniques should be employed to avoid such contamination. Proteus is a group of rod-shaped pathogenic bacteria, family Enterbacteriaceae, which is associated with food borne illnesses [19,20]. In the present study, Proteus was isolated from contaminated fish and consumption without proper cooking may lead to a disease known as scombroid poisoning. Bacterial isolates with methicillin resistant genes were detected in bacterial strains isolated from sewage water collected from fish markets, animal slaughter house and community sewage [13]. Fishes might have contaminated through poor hygiene or improper handling. Motile aeromonads are more common in marine fish samples. Aeromonas species were isolated from the samples which are very common bacterial pathogen found in fish samples. It was reported to produce aerolysin toxin [21]. Presence of such pathogenic strains of bacteria causes cross contamination in sea foods which can create risk of infection in immuno-compromised patients and children.
Fish products were often found to contain large numbers of faecal coliforms, particularly Enterococcus species [22]. The most dominant bacterial species in fish samples were Enterococcus species, an opportunistic pathogen commonly found in fish samples. However, the presence of Streptococcus was less when compared to Enterococcus species. Streptococcus were also isolated from the fishes which are an opportunistic pathogen and in unusual circumstances it causes infection in human. The presence of bacteria in the fish sample indicates the general quality of the fishes or the degree of spoilage. It also resembles the hygienic status of the food product and storage conditions. It becomes really a concern when the human population uses undercooked or under processed fishes. There should be a continuous observation to detect the pollution of food products and thereby to control pathogens transferring through food products.
Antibiotic resistance is becoming common where the usage is uncontrolled especially in aquatic environments. Antibiotic resistance occurs in aquatic environment rapidly as fast as less than one minute by the transfer of R plasmid [23]. In the present study, 38.4% of Proteus species showed MDR. The development of resistance in Proteus mirabilis with chromosomal AmpC-type with beta-lactamase had been reported [20]. Bacteria showing resistance towards four and above are considered as MDR strains [22]. Some of the bacterial isolates showed antibiotic resistant pattern to amoxicillin and gentamycin and all the strains were resistant to bacitracin. High rates of resistance in E. coli isolate to antibiotics such as erythromycin, amoxicillin and tetracycline has reported earlier in clinical samples [24]. Similarly 77.4% MDR E.coli isolated from poultry workers, chicken meat samples in Saudi Arabia [25]. In the present study, 40% of the E. coli isolated showed MDR and the MAR index was 0.44. Aeromonas and Enterobacter species showed MDR and the MAR index was 0.36 and 0.35 respectively. High level of antibiotic resistance with ampicillin (85.5%), rifampin (57.1%), streptomycin (49.1%) and nalidixic acid (44.6%) in Aeromonas species isolated from farmed fresh water animals were reported earlier [26]. Enterococcus was the major strain (n=16) isolated from fish samples and four (25%) of these bacteria showed MDR. Enterococcus has been reported for resistance by various mechanisms including altering the drug targets, inactivating the drug, through efflux pumps, modification of cell envelope etc., [27]. The MAR index of Enterococcus species was 0.39 which was categorised under high risk contamination. Streptococcus species (25%) were resistant to four antibiotics and the MAR index was 0.33. Recent studies showed that Streptococcus is emerging with antibiotic resistance towards cefotaxime, clindamycin, ceftriaxone, erythromycin, levofloxacin and tetracycline [28]. Since it is resistant to bacitracin, it cannot be grouped under highly pathogenic strains of Group A Streptococci. Carbapenem resistance gene has been found in bacteria isolated from municipal sewage collected from Saudi Arabia [29]. In the present condition, the uncategorised use of antibiotics in human, animals and aquatic environment leads to the formation of antibiotic resistant pathogens which are becoming a threat leading to more stress and health risk on the public [30]. In the present study, the occurrence of MDR bacteria has been identified in fish samples collected from local market. It may become serious threat to the public when these food products are consumed by human thereby leading to infection by MDR bacteria.
Limitation(s)
The study was carried out for three months. Similar studies with large number of samples and wide geographical location can draw a final conclusion on the source of contamination and seriousness of the threat of antibiotic resistance. Further identification of resistance genes in the bacterial isolates can also limit the spread of MDR bacteria.
Conclusion(s)
Proper hygiene and modern technologies should be practiced to avoid cross contamination of fishes in the local market. Molecular level studies are needed to detect the ways of contamination, gene transfer or mode of acquiring resistance in bacteria. The usage of high dose of preservatives and antibacterial agents in the fishes during storage can be avoided to eliminate MDR strains. Proper surveillance and implementing appropriate measures can control spreading antibiotic resistant bacterial strains through seafood.
AMX: Amoxcillin (10 μg); B: Bacitracin (10 μg); C: Chloramphenicol (30 μg); CIP: Ciprofloxacin (5 μg); E: Erythromycin (15 μg); GEN: Gentamycin (10 μg); TE: Tetracycline (30 μg)