The enzyme 5,10-MTHFR catalyses the reduction of 5,10 methylene Tetrahydrofolate (THF) to 5-methyl THF (5-MTHF). The essentiality of 5-MTHF in methionine cycle and S-Adenosyl Methionine (SAM) formation is not unknown. SAM in turn acts as donor of methyl group that is required for various metabolically demanding trans-methylation reactions such as synthesis of creatine from guanidoacetate, phosphotidylcholine from phosphatidylethanolamine, epinephrine from nor-epinephrine, choline from ethanolamine, melatonin from acetyl serotonin, methylation of Deoxyribonucleic Acid (DNA) [7]. The metabolic implications of trans-methylation reactions pertain to biosynthesis of these active endogenous compounds are essentially required for modulating the functionalities of protein and nucleic acid involved in regulation of gene expression like DNA methylation is implicated in gene expression and genome imprinting. Such modifications allow controlled expression of developmental genes during the embryonic period [8,9].
Methionine cycle utilises 5-MTHF for regeneration of methionine from SAM through homocysteine. Reduced bioavailability of 5-MTHF would lead to hyperhomocysteinemia that is accounted for prothrombotic effects by endothelial dysfunction, activation of platelets and fibrin formation and inhibition of natural anticoagulants resulting in venous thrombo-embolic phenomenon like stroke, repeated pregnancy loss, vaso-occlusive crisis in haemoglobinopathies like Sickle Cell Disease (SCD) and thalassaemia, neural tube defects in fetus and also cancer [10,11].
Human MTHFR gene is mapped at 1p36.3 with 11 exons. Over 40 polymorphisms viewed in various studies, two SNPs, C677T (exon 4) and A1298C (exon 7) have been implied for various clinical presentations. C677T is the result of replacement of cytosine by thymine at 677th position of gene sequence and substitutes alanine to valine at 22nd amino acid at the protein level [12]. This makes the enzyme thermolabile. The enzyme activity is reduced by 35% in heterozygote (CT) genotype and by 70% in homozygous (TT) genotype [13].
These polymorphisms depict heterogenous distribution globally. The frequency of the two alleles in Indian population is reported to be 10.08% and 20.66% for MTHFR 677T and 1298C respectively [14]. The homozygous variant form, 677T, is reported to be more prevalent in North Indian population whereas 1298C is more prevalent in East and South India [15]. The SNPs distribution of MTHFR in the region of Central India is not much studied, especially in children. It is of utmost importance to know the burden of SNP in children so that therapeutic modulation may be re-considered at an early age in disorders associated with the said polymorphisms. For example, the allelic and genotype distribution of C677T and A1298C polymorphisms in this state will enable to re-think regarding the intervention protocol like folate supplementation in children with vascular diseases like sickle cell anaemia. Therefore, the study was aimed primarily to know the prevalence of MTHFR variants in children in this region.
Materials and Methods
The cross-sectional study was conducted on 375 children aged 5-15 years of age. All children in the study population, irrespective of their health status, were recruited from Out-Patient Department (OPD) and In-Patient Department (IPD) of the institute during the sampling period of three months, May 2018 to July 2018.
The study was approved by the Institute Ethics Committee (AIIMSRPR/IEC/2016/020) and written informed consent was signed by the parents/guardians/legal representatives before enrollment.
Under all aseptic conditions, 3 mL of blood samples were collected in Ethylenediaminetetraacetic Acid (EDTA) vacutainers. The samples were processed for DNA extraction within two hours of collection. The DNA from the whole blood was extracted using commercially available kit InvitrogenTM PureLinkTM Genomic DNA Mini Kit from ThermoFisher Scientific (Catalog number K182002). The protocol for extraction was followed according to manufacturer’s instructions provided along with the kit [16,17]. Both the polymorphism along with allelic discrimination were genotyped by pre-validated TaqMan based SNP Genotyping real-Time Polymerase Chain Reaction (PCR) Assays (Applied Biosystems, ThermoFisher, Foster City, CA), C677 (rs1801133) [18] and A1298 (rs1801131) [19] processed on CFX96 real-Time PCR system, Biorad (USA). Each pre-formulated assay contained two unlabelled PCR primers, forward and reverse primers, one each of Applied BiosystemsTM VICTM and 6FAMTM dye-MGB labeled probe to detect both alleles.
Statistical Analysis
Percentage calculation for allele and genotype frequency, HWE and LD were calculated using Microsoft Excel. For all statistical calculations, p-value less than 0.05 were considered significant.
In the text below, C´ would be used with intention to differentiate the C allele of A1298C with that of C677T wherever applicable. The same would be used henceforth in the below manuscript.
The genotypic frequency is the number of its occurrence (n) in the study population whereas the allelic frequency was calculated as the ratio of the total number of occurrence (n) to the total number of alleles (double the sample size=750). The Minor Allele Frequency (MAF) was used to depict the frequency of occurrence of the mutant alleles, T allele for C677T and C´ allele for A1298C SNPs [20].
The genotype and allelic frequencies of the two polymorphisms were tested for HWE using chi-square test [21]. To assess the HWE, the expected and observed number of subjects were calculated. The expected number of subjects with a specified SNP genotype was calculated by multiplying the frequency of that genotype as shown in [Table/Fig-1], with the total sample size (n=375) e.g., the expected number of subjects with CC genotype was 0.86*0.86*375=277. The statistical deviation from the old equilibrium can be derived by using simple Chi-square test. Chi-square statistics for the observed and expected genotypic frequencies was calculated for HWE.
The frequency of occurrence of MTHFR genotypes for both polymorphisms in the population when conformed to Hardy-Weinberg equilibrium.
| C (0.86) | T (0.14) | | A(0.59) | C´ (0.41) |
|---|
| C (0.86) | CC (C2)(0.74) | CT (0.12) | A (0.59) | AA (A2)(0.35) | AC´ (0.24) |
| T (0.14) | CT (0.12) | TT (T2)(0.02) | C´ (0.41) | AC´ (0.24) | C´C´ (C´2)(0.17) |
The Hardy-Weinberg equation, p2+2pq+q2=1, is a mathematical equation used to estimate the frequency of genotypes in a population at genetic equilibrium. Considering the C and A as dominant allele and T and C´ as mutant recessive allele, at genetic equilibrium indicated the frequencies of three genotypes as homozygous wild type (p2), heterozygous variant (2pq) and homozygous mutant type (q2) [22].
The observed (Oi) and expected (Ei) number of subjects with each haplotypes, CA, TA, CC´ and TC´, were calculated as discussed before for the haplotype frequency table. Chi-square statistics for the observed and expected haplotype frequencies were calculated to assess the LD between the two SNP loci.
The LD between the polymorphisms was assessed along with haplotype association analysis in order to predict the non-random association of the alleles in the population [23]. The LD coefficient (D) between the two SNPs was calculated using the formula D=(PCA*PTC´) – (PCC´*PTA). The calculated D (-0.013) in this population was found to be less than zero. In a biallelic system, the actual gamete frequencies cannot be negative. Hence, Lewontin’s coefficient or ratio (D´) was applied for standardisation of the differing allele frequencies. As the observed D was found to be negative, D´ was calculated as D/Dmin that implied the ratio of D to its maximum possible frequencies of dominant alleles (PA*PC). Hence, D´=0.013/0.51=0.025 [24,25].
Pearson’s correlation coefficient (r2)=D2/PCPTPAPC´=0.0056 was also calculated to measure the LD. To check the significance of LD between the alleles, χ2 was calculated as r2N, where N is the total number of alleles [24,25].
Results
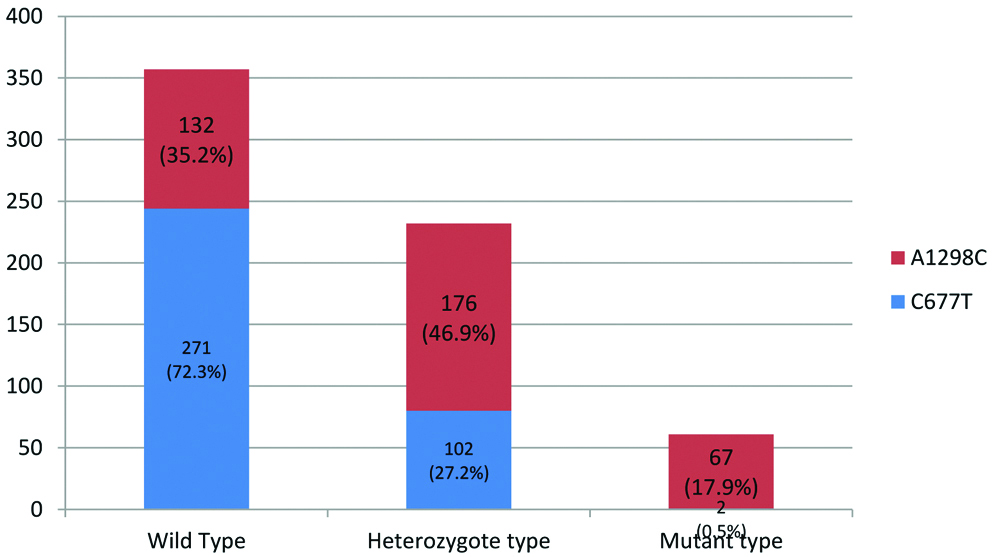
The [Table/Fig-2], indicated that the most occurring genotype in the study group was wild type CC677 (n=271) followed by heterozygous A1298C (n=176). Individuals with heterozygous and homozygous mutant variants of A1298C were observed to be more prevalent than that of C677T variants.
Distribution of MTHFR genotypes in the study population (n=375).
As depicted in the [Table/Fig-3], wild type C allele of C677T was the commonest of all alleles in the study population. The MAF (C´) of A1298C is nearly three times to that of the T allele of C677T.
The allelic frequency of the two SNPs in the total allelic population (n=750).
| A1298C |
|---|
| A allele | C´ allele |
| 440 (0.59) | 310 (0.41) |
| C677T |
| C allele | T allele |
| 644 (0.86) | 106 (0.14) |
As enumerated in [Table/Fig-4], the highest frequency was observed for CC/AC´ whereas the lowest was for TT/AA. No children depicted TT/AC´ and TT/C´C´ combinations.
Distribution of combined genotype in the study population (n=375).
| Different combined genotypes N (%) |
|---|
| 1 | CT/AA | 53 (14.1%) |
| 2 | TT/AA | 02 (0.5%) |
| 3 | CC/AA | 77 (20.5%) |
| 4 | CT/AC´ | 44 (11.7%) |
| 5 | TT/AC´ | 0 |
| 6 | CC/AC´ | 132 (35.2%) |
| 7 | CT/C´C´ | 05 (1.3%) |
| 8 | TT/C´C´ | 0 |
| 9 | CC/C´C´ | 62 (16.5%) |
As shown in the [Table/Fig-5], the observed frequency for the heterozygous CT variant increased by 13% whereas TT variant of C677T was reduced by 71%. The heterozygous AC´ of A1298C was increased by 1.8 times.
Comparison for the expected and observed frequency for the MTHFR variants in the study population (n=375).
| Expected number of subjects | Observed number of subjects |
|---|
| CC | 277 | 271 |
| CT | 90 | 102 |
| TT | 07 | 02 |
| AA | 130 | 132 |
| AC´ | 63 | 176 |
| C´C´ | 91 | 67 |
Hardy-Weinberg equation was applied to find out the equilibrium for a gene and the allelic frequencies. The distribution of C677T genotypes followed HWE and did not vary significantly from that of expected frequency. However, the observed frequency of MTHFR A1298C polymorphisms did not follow HWE. The number of children with A1298C genotype was significantly more than expected (176 vs 63, χ2=45.38, p<0.001). The frequency of heterozygous and homozygous mutant variant of A1298C in this region was observed to be more prevalent as compared to C677T. This could be ascribed to the hypothesis that the mutation is disrupting the equilibrium of allele frequencies of A1298C SNP by introducing the mutant C´ allele into the population.
The haplotype frequency table [Table/Fig-6] confers that the dominant haplotype in the population was 677C-1298A (0.51) followed by 677C-1298 C´ (0.35).
Haplotype frequency table.
| A1298C |
|---|
| C677T | | A (0.59) | C´ (0.41) |
| C (0.86) | CA (0.51) | C C´ (0.35) |
| T (0.14) | TA (0.08) | T C´ (0.06) |
In order to assess the LD, χ2 was calculated for the observed and expected frequencies of the haplotypes as shown in the [Table/Fig-7]. The χ2 was 16.03, it was found to be significant (p<0.001). This states the rejection of linkage equilibrium between the two SNPs. The measure of disequilibrium, D≠0, that denoted that the expected frequency was not met in the study population. In order to nullify the negative value of D (-0.013), Lewontin’s coefficient (D´) was applied. The D´ was found to be 2.5% that reflected the disequilibrium upto 2.5% of the theoretical maximum of the given alleles.
Comparison for the expected and observed frequencies of the haplotypes.
| Haplotype | Observed number of subjects | Expected number of subjects |
|---|
| CA | 306 | 381 |
| TA | 99 | 62 |
| CC´ | 243 | 264 |
| TC´ | 49 | 43 |
The Pearson’s correlation coefficient (r2) between the pair of SNPs was also used as a measure of LD which was calculated to be 0.006. The χ2=r2N distributed with df=1 was calculated to be 4.2 which was significant (p=0.04).
Discussion
Optimal activity of the enzyme MTHFR is critical for maintaining the cellular methionine-homocysteine cycle. Genetic variants of the enzyme adversely influence the vascular events in various disorders. The heterogenicity of these polymorphisms in different geographical regions mandated the need to understand its distribution in our region as well. The primary objective of the study was to determine the frequency of MTHFR C677T and A1298C SNPs in this region.
As depicted in [Table/Fig-2], the wild type CC677 (72.3%) was the most occurring genotype in the study group. The heterozygous (46.9%) and homozygous mutant (17.9%) of A1298C were the more frequently occurring than that of C677T (27.2%) and 677TT (0.5%) variants.
Saraswathy KN et al., in their study in 23 different Indian populations from five geographical regions, tabulated a highest genotypic frequency of 82.2% for CC677. The frequencies of AC´ and C´C´ genotypes were 18.5% and 11.4% respectively whereas CT and TT were 15.27% and 2.45% respectively [15]. The study by Rai AK et al., in 165 Indian mothers of control group, frequency of CC was 75.1% followed by 52.8% for AC´, 40% for AA, 23.6% for CT, 7.1% for C´C´ and 1.2% for TT [26]. The genotype frequencies observed in present study was almost similar to the findings of Rai AK et al., [26].
The frequency of wild type alleles, A and C were 0.59 and 0.86 respectively [Table/Fig-3]. The frequency distribution was found to be nearly similar to the observations reported by Sukla KK and Raman R with a frequency of 0.69 and 0.89 respectively [20]. The present study findings were found to be close to allelic frequencies, 0.66 (for allele A) and 0.87 (for allele C), reported by Rai AK et al., [26].
The Minor Allele Frequency (MAF) for A1298C was 0.41 and that of C677T was 0.14 in the study population [Table/Fig-3] that confirms the higher prevalence of mutant C´ allele of A1298C. The observations nearly corroborated to the frequency of 0.31 and 0.11 respectively for T and C´ alleles as reported by Sukla KK and Raman R and also to Rai AK et al., with frequencies of 0.12 and 0.33 respectively [20,26]. The MAF also corroborated with T and C´ frequencies of 0.41 and 0.1 respectively in 420 South Indian adult population as published by Devi ARR et al., [27]. Saraswathy KN et al., in their study depicted a frequency of 0.1 for T allele and 0.2 for C´ allele [15]. The occurrence of mutant C´ allele was nearly doubled in our population. Hence, the frequency of heterozygous A1298C (46.9%) was found to be more in the present study population. As a reason, the most prevalent combined genotype is CC/AC´ (35.2%) (as shown in [Table/Fig-4]).
This could be explained by the fact that the gene and the C´ allelic frequencies did not follow HWE [Table/Fig-5]. HWE refers to a neutral equilibrium that describes the distribution of genotype frequencies in a population. The theorem assumes that, in a non-evolving population of diploid, sexually reproducing individuals, the allele frequencies in that population would not change from generation to generation. However, if the allele frequency changes, either due to mutation or non-random mating or genetic drift due to very large population size, a new equilibrium will be observed [21,22]. Hence, it is reasonable to hypothesise that the mutation is disrupting the equilibrium of allele frequencies more so in A1298C than C677T by introducing the mutant C´ allele into the population. Hence, the frequency of heterozygous and homozygous mutant variant of A1298C in this region was observed to be more prevalent as compared to C677T. The observed genotype frequencies of MTHFR C677T and A1298C were found to follow HWE in Chinese population as decoded by Yang B et al., Wang X et al., and Fan S et al., [21,22,28]. Similarly, Murthy J et al., in their study also revealed a genetic distribution of the MTHFR genotypes very much similar to the expected distribution under HWE [29].
The nearby states West Bengal and Maharashtra also deferred prevalence of 30% and 26% respectively for mutant 1298CC as observed by Kumar J et al., [30]. In a study conducted by Nishank SS et al., on 150 SCD cases and 150 control subjects in Central India recorded a genotype frequency of 28% and 14.6% for heterozygous and mutant variants of A1298C [31].
The allelic frequencies observed in this study also corroborated with the MAF of 0.15 for C677T and 0.43 for A1298C published by Kumar J et al., in 203 cases of coronary artery disease in North India [30]. The distribution of the two MTHFR polymorphisms in Indian population with 834 C677T alleles and 894 A1298C alleles reported MAF of 0.129 and 0.38 respectively and homozygous mutant genotype frequency of 2.16% (677TT) and 19.46% (1298CC). The high prevalence of 677TT mutant variant were mostly observed in Northern states like Himachal Pradesh (9.75%) and Punjab (6.12%) and Southern part of India like Andhra Pradesh (5.56%) [30]. The Central India reported no mutant variant for this SNP. This could be the reason for low prevalence of 677TT mutant genotype in the present study population. The recorded frequency of mutant 677TT genotype in Indian population is much lower as compared to Chinese (16.9%) and Japanese (15.6%) whereas the mutant homozygous 1298CC variant is much more prevalent in India as compared to China (3.3%) and Japan (1.6%) as reported by Shrubsole M et al., and Hiraoka M et al., respectively [32,33]. The geographical and ethnic variations in the frequencies of C677T and A1298C observed by Wang X et al., in their study might justify the distribution pattern observed in the present study [22].
T allele prevalence is not common in African (10.3%) and Asian population (19.7%) but is found to be more in European population (34.1%) [34]. Balasa VV et al., reported a prevalence of homozygosity for C677T MTHFR variant to be 5% in 40 homozygous SCD cases in Ohio, USA [35]. This could be due to the high prevalence of homozygous C677T mutant variant in US population as reported by Botto LD and Yang Q [36].
The haplotype frequency [Table/Fig-6] depicted the dominant haplotype to be 677C-1298A (0.51) followed by 677C-1298 C´ (0.35), 677T-1298A (0.08) and 677T-1298 C´ (0.06). In a study conducted by Abu-Hassan DW et al., on 264 females conformed that the most prevalent haplotype was 677C-1298A (35.4-51.5%) followed by 677C-1298C´ (14.4-39.0%) [37]. However, the haplotype frequency 677T-1298A (43.6%) was most prevalent than 677C-1298A (37.9%) in Chinese population as revealed by Fan S et al., [28].
The distribution of haplotype frequencies differed significantly (p<0.001) from the expected frequencies as depicted in [Table/Fig-7]. This referred to LD between the two loci of the MTHFR SNPs in the study population. When the coefficient of LD, D=0, the alleles are said to be in linkage equilibrium, which means they occur randomly and not co-inherited. When D≠0, it implied that at least one of the observed haplotype was not observed as expected. The negative value of D (D=-0.013) indicated that a negative LD existed i.e., C allele (dominant) of locus C677T is linked with the C´ allele (mutant) of A1298C locus. The Lewontin’s coefficient, D´ was found to be 0.025 that reflected the disequilibrium upto 2.5% of the theoretical maximum of the given alleles.
Significantly low r2 (0.006) reflected the low allele frequency. This concluded that the alleles exhibited non-random association and one of the SNPs (677TT) depicted a very low allele frequency. The alleles that did not follow HWE might be accounted for the low LD between the two polymorphisms. Such LD was also revealed by Fan S et al., and Abu-Hassan DW et al., in their respective studies [28,37].
Callejon G et al., in their study disclaimed the LD between the alleles of the C677T and A1298C polymorphisms in 342 foetal samples [38]. Similarly in a study conducted on 325 subjects in North Indian population also observed low TT genotype, the population not in HWE and medium range LD (D’=0.68, p<0.001) [39].
Homozygous form for both mutants (TTC´C´) was not found in the study. As suggested by Callejon G et al., genotype of four mutations might be considered as non-viable as none of the 468 live foetus had such genotype. This could be due to the fact that the said combination of homozygous variant must be under stringent gene selection process as the homozygous mutant variant of both polymorphisms result in severe sequel like non-viability of the foetus resulting in abortions [38].
Limitation
The major limitations in the study were the small sample size and non-inclusion of any disease cases like sickle cell anemia, to observe the impact of the polymorphism in cases.
Conclusion
The mutant allele of A1298C was more frequently occurring and thus the heterozygous and homogygous mutant variants of A1298C were more prevalent than C677T variants. The hypothesis that incorporation of mutation in A1298C is likely contributing the mutant allele into the population and thereby accounts for disruption of HWE and low LD between the two polymorphisms.
The baseline data observed in this study would open perspectives for future research in this field. The mutant allele gradient in this geographical region could be the influence of various environmental factors on gene expressions. The evidence based genomic data observed in this region could be the foundation for further studies on gene-mapping and environmental-genetic interactions with clinical implications in various diseases.
Recommendations
Further studies on the mutation rate, the gene flow, the genetic drift, recombination and mating population on large scale population is required to claim regarding LD between the two MTHFR polymorphisms.