Fluoroquinolones are a group of synthetic drugs with an antibacterial activity that is widely used in the treatment of bacterial infections, both gram-negative and gram-positive bacteria [1]. Therefore, new derivatives of fluoroquinolones may also have a reverse effect on bacterial replication. In the base structure of quinolones, the Carboxyl group at position 3 and Ketone at position 4 are essential for antibacterial activity. The substitution of a Fluorine atom at position 6 is a way of distinguishing fluoroquinolones from quinolones, and it is a factor in increasing the effect on gram-positive and gram-negative bacteria. All the compounds present in the antibiotic group of fluoroquinolones are similar in terms of the mechanism of action and stop DNA replication in susceptible bacteria selectively and reversibly [2].
DNA gyrase is an enzyme essential for the survival and proliferation of gram-positive and gram-negative bacteria, but not present in eukaryotes [3]. The two strands of DNA are in a double helix in a cell, but they must be separated from each other in the process of replication and transcription. To facilitate the opening of the double-stranded DNA, the DNA gyrase enzyme relieves the strain while the double-stranded DNA is being unwound [4]. This enzyme is an attractive target for antibacterial drugs [4]. The DNA gyrase is an enzyme within the class of topoisomerase consisting of two A and B subunits. The fluoroquinolones inhibit the GyrA subunit from the Types II and IV topoisomerase enzymes and form the relaxation complex analogue that prevents the DNA unwinding during replication [5].
Therefore, fluoroquinolones cause bacterial death by inhibiting DNA replication and transcription. Norfloxacin and ciprofloxacin are among the first synthetic compounds of fluoroquinolones that have antibiotic dose-dependent bactericidal activity [6]. Fluorination of quinolones enhances drug penetration into bacterial cells and increases their efficiency [7].
The addition of the cyclopropyl group at position 1 (enrofloxacin and ciprofloxacin), ethyl or fluorophenyl (difloxacin, levofloxacin) improves the antibacterial activity spectrum on gram-negative and gram-positive bacteria. Adding the piperazine group at position 7 to the core of the fluoroquinolones improves their activity on Pseudomonas aeruginosa and other gram-negative bacteria [8]. Additionally, changing the nitrogen group at position 8 to the carbon group reduces the negative effects of these compounds on the central nervous system. Therefore, any brief changes in the structure of the fluoroquinolones are responsible for the development of various physiological properties, antibacterial strength, and spectrum of these drugs [9].
The synthesis and application of organo-fluorine compounds are an important part of medical and therapeutic research. These compounds are widely used in the pharmaceutical industry, chemistry, and materials as well as in agriculture. The importance of organofluorine compounds are due to the unique biological properties of their fluorine atoms [7]. Trifluoromethyl compounds are also effective because they have high electron acceptor properties, and give them unique lipophilic properties. This feature facilitates drug penetration into the microbial cell membrane and thus increases the drug half-life in the body.
In recent years, a large percentage of bacteria due to conferring genes and mutational alterations have become resistant to fluoroquinolones. Therefore, we should look for new derivatives of these antibiotics [10-12]. Some of the new compounds can act against quinolone-resistant mutants, and against the targets, namely the subunit A of the DNA gyrase enzyme, as well as topoisomerase IV of prokaryotes [13].
In the most important drugs which are commercially available, for example, antidepressant fluoxetine, cholesterol-lowering drug atorvastatin and the antibacterial ciprofloxacin at least one fluorine atom are present. The trifluoromethyl group is becoming more and more important in both of agrochemical and pharmaceutical applications because of the influence of fluorine atom on physical, chemical, and physiological properties, stability, and lipophilicity of the molecule. In recent years, reactions of trifluoromethylation have been extensively investigated, in order to synthesise new biologically active organofluorine compounds. These synthesised compounds have quinolone base structure which are trifluoromethylated by trifluoroacetimidoyl chlorides in pyrazine ring. Therefore combination of trifluoromethyl group and quinolone framework increase the biological activity of compounds [11-13].
The current study examined the inhibitory effect of two compounds derived from fluoroquinolones; N-4-methyl (phenyl) -2,2,2-trifluoroacetimidoyl ciprofloxacin (C1) and N-4-methyl (phenyl) -2,2,2-trifluoroacetimidoyl norfloxacin (C2), on S. aureus and E. faecalis, which are clinically important and resistant to a wide range of antibiotics in terms of inhibitory growth potency and expression level of DNA gyrase genes.
Materials and Methods
This experimental in-vitro study examined the antimicrobial effects of two synthetic compounds including N-4-methyl (phenyl) -2,2,2-trifluoroacetimidoyl ciprofloxacin (C1), and N-4-methyl (phenyl) -2,2,2 trifluoroacetimidoyl norfloxacin (C2) from June 2018 to November 2018. Norfloxacin and ciprofloxacin (Sigma-the USA) were used as the base molecules to synthesise mentioned compounds. These compounds were synthesised in the Chemistry Department, Faculty of Sciences, Vali-e-Asr University of Rafsanjan, Iran. Also, the compounds examined for antimicrobial properties and, molecular test in Rafsanjan University of Medical Sciences, Kerman, Iran. This study was conducted in accordance with the institutional ethics code No: IR.RUMS.REC.1395.139.
Synthesis of Fluoroquinolones-Derived Compounds
Synthesis of C1 (N-4-methyl (phenyl)-2,2,2- trifluoroacetimidoyl ciprofloxacin) and C2 (N-4- methyl (phenyl)-2,2,2-trifluoroacetimidoyl norfloxacin) has been described in the previous study by the present authors and the structure of synthesis compounds (C1 and C2) have been identified by FT-IR, 1H-NMR, 13C-NMR spectroscopy analysis and elementally analysis according to the literature reported [11,12].
Cultivation and Determination of Bacterial Sensitivity to Synthetic Compounds
S. aureus PTCC 1431 (ATCC 25923) and E. faecalis PTCC 1393 (ATCC 11700) were obtained from Persian Type Culture Collection and removed from lyophilization state by adding the normal saline, according to the manufacturer’s instructions, and then cultured in Blood Agar (BA, Merck) to revive, followed by purification. Using the BMD protocol in the Muller Hinton Broth (MHB, Merck), Minimum Inhibitory Concentration (MIC) was calculated for each of the synthesised compounds as follows: 1024 μg/mL of each of the above compounds was taken in order to perform the doubling dilution step down technique. An appropriate amount of bacterial suspension of 0.5 McFarland turbidity standard prepared in the MHB medium was taken and added to each well so that the bacterial concentration was 104 CFU/mL per well.
The MIC was determined for each of the compounds and each bacterium based on the lack of growth in the last well [14]. In the disc diffusion technique (Kirby-Bauer Test), seriate dilutions of 1:2, 1:4, 1:8, 1:16, 1:32 1:64, 1:128, 1:256 and 1:512 were prepared from the initial concentration (1024 μg /mL) of the compounds C1/C2 to compare the mean diameter of the Zone of Inhibition (ZOI) on tested bacteria. The ZOI was measured around antimicrobial-containing discs based on MIC value obtained from the previous test using the BMD. An appropriate amount of bacterial suspension of 0.5 McFarland turbidity standards was taken and cultured by swab on Muller Hinton Agar (MHA, Merck). Next, the discs containing the specified amount (MICs were obtained from the previous test) of synthetic antimicrobials were cultured on the MHA medium. The culture plates were incubated at 37°C for 24 hours and then ZOIs were measured. It should be noted that vancomycin (V30 μg/disc, PadtanTeb, Iran), was used as a positive control in the form of powder and discs, BMD and disk diffusion tests. To increase the accuracy of the results for the disc diffusion method, the tests were performed in three replicates.
Evaluation of DNA Gyrase Gene Expression
At first, the fresh culture of S. aureus and E. faecalis bacteria was prepared on BA. The colonies of these bacteria were then used to inoculate onto Tryptic Soy Broth (TSB, Merck). After that, this broth media were separately treated by the 1.2×MIC concentration of C1/C2 and then incubated and centrifuged at 7°C with 180 rpm. The Optical Density (OD) of the samples was read at a wavelength of 650 nm, with the OD value of 0.6 for all samples [15].
Total mRNA was extracted from broth media containing bacteria and treated with C1/C2 by RNA extraction kit (QIAGEN Ltd., USA) according to the manufacturer’s instructions. To remove the genomic DNA, the extracted samples were treated with a DNAse prepared in the kit. After extracting mRNA, the purity was determined by spectrophotometry at wavelengths of 260 and 280 nm. The ratio of these two wavelengths was between 1.7 and 1.9 for more than 80% of the extracted samples. In order to the synthesis of cDNA from extracted total mRNA, cDNA synthesis kit (Takara, Japan) was used and according to the manufacturer’s instructions, the extracted mRNAs were converted to cDNA using random hexamer and oligo-dT primers as well as reverse transcriptase enzyme. The real-time PCR assay was performed to investigate the changes in DNA gyrase gene expression using specific primers of DNA gyrase as the target gene and 16sRNA as a reference gene using SyberGreen method and ABI Step One Plus (Applied Biosystems, USA).
Statistical Analysis
The obtained data were analysed by ABI version 2.3 software by the equation of 2 –ΔΔCt formula. The sequence of primers used in the study is specified in [Table/Fig-1]. Data collected from evaluating antibiogram and BMD tests and measuring the ZOIs diameters [Table/Fig-2,3], and the results of gene expression (using real-time PCR) were analysed by SPSS version 18.0 software. “One-way ANOVA” was used to analyse the data and “Tukey’s multiple comparison test” was used if significant. The results of the statistical analysis were reported as mean±standard deviation, and the significance level in the tests was considered as 0.05.
Demonstrates the sequences of primers used in the study.
| Gene | Forward | Reverse |
|---|
| DNA gyrase subunit A | TGGCCCAAGACTTTAGTTATCGTTATCC | TGGGGAGGAATATTTGTAGCCATACCTAC |
| 16s RNA | TGT CGT GAG ATG TTG GG | CGA TTC CAG CTT CAT GT |
Minimum Inhibitory Concentrations (MICs) of synthesised compounds compared to an appropriate antibiotic against Staphylococcus aureus and Enterococcus faecalis (μg/mL).
| Bacterial species | Compound | MIC (μg/mL) |
|---|
| 512 | 256 | 128 | 64 | 32 | 16 | 8 | 4 |
|---|
| Staphylococcus aureus | C1 | − | − | − | − | − | + | + | + |
| C2 | − | − | − | − | − | − | + | + |
| V | − | − | − | − | − | − | - | + |
| Enterococcus faecalis | C1 | − | − | + | + | + | + | + | + |
| C2 | − | − | − | + | + | + | + | + |
| V | − | − | − | − | − | − | + | + |
(+): No inhibition (Bacterial growth not affected); (-): Inhibition (Bacterial growth affected)
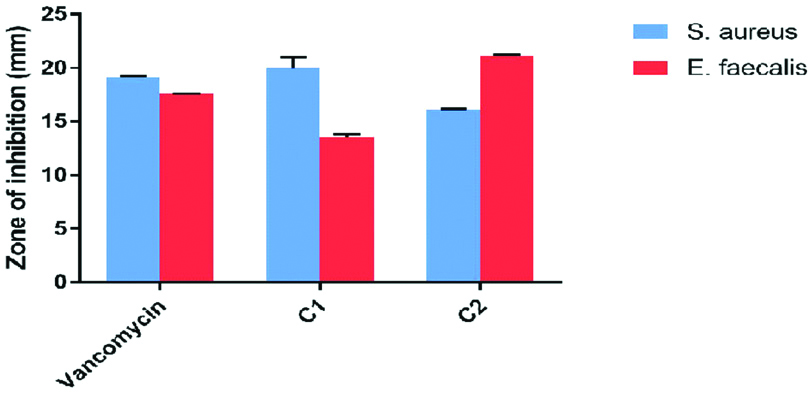
Inhibition zone caused by antibacterial candidates and therapeutic antibiotic against Staphylococcus aureus and Enterococcus faecalis (Triple tests).
| Compound | Inhibition zone (mm) |
|---|
| Gram-positive bacteria |
|---|
| Staphylococcus aureus | Enterococcus faecalis |
|---|
| C1 | (32 μg) 20±0.2 | (256 μg) 13.5±0.1 |
| C2 | (16 μg) 16.1±0.1 | (128 μg) 21.1±0.2 |
| Vancomycin (30 μg) | 19.1±0.2 | 17.5±0.1 |
Results
Antimicrobial Activity
The results of this study showed that the MIC value of C1 for S. aureus was 1:32, and in dilution of 1:16 for C2. The inhibitory effect of vancomycin as a standard antibiotic was found 1:8, indicating a higher inhibitory effect of vancomycin against S. aureus than two synthetic compounds [Table/Fig-2]. The MIC values for the compounds C1/C2 regarding E. faecalis were 1:256 and 1:128, respectively. The vancomycin was able to prevent the growth of E. faecalis in the dilution of 1:16, and hence the inhibitory potency of vancomycin was higher than that of the two tested synthetic compounds. Overall, the growth-inhibitory potency of the C1 was less than the C2 [Table/Fig-2].
The results of the disc diffusion method showed that the mean ZOI diameters induced by C1 and vancomycin on S. aureus were 20.0±0.2 mm and 19.0±0.1 mm, respectively and were not statistically significant (p=0.14), while in the presence of C1 and vancomycin, the mean ZOI diameters on E. faecalis were 13.5±0.1 mm and 17.5±0.1 mm, respectively that was statistically significant (p=0.01). Therefore, vancomycin had a more inhibitory effect, especially on E. faecalis, in comparison to C1. The results showed the mean ZOI diameters of C2 and vancomycin on S. aureus were 16.1±0.1 mm and 19.1±0.2 mm, respectively (p=0.01), whereas the mean ZOI diameter of C2 and vancomycin on E. faecalis were 21.1±0.2 mm and 17.5±0.1 mm, respectively (p=0.01). Therefore, in comparison with vancomycin, the C1 had a less effect on S. aureus while the C2 had higher antibacterial activity against E. faecalis. [Table/Fig-3,4].
Zones of Inhibition of Vancomycin, C1 and C2 on S. aureus and E. faecalis.
DNA Gyrase Gene Expression
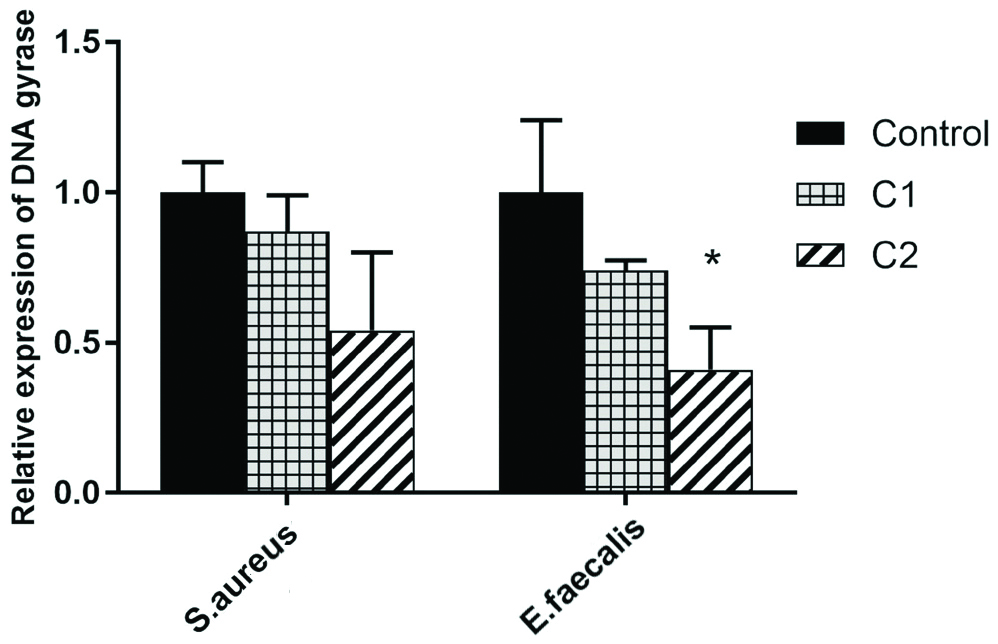
The results of the gene expression showed that C1 could reduce the DNA gyrase gene expression in S. aureus and E. faecalis, but not statistically significant (p=0.06). Compound C2 reduced the DNA gyrase gene expression in both bacteria; this decrease was statistically significant for E. faecalis (p<0.01) and used control was anti-bacterial free medium [Table/Fig-5].
Demonstrates alteration in DNA gyrase at mRNA level, after treatment with C1 and C2 of S. aureus and E. faecalis. Data are presented as mean±SD, *significant (p<0.05).
Discussion
The present study showed that the vancomycin has a higher antimicrobial effect in comparison with the C1, especially on E. faecalis, and probably C1 is unable to overtake vancomycin regarding gram-positive bacteria. The effect of C2 on S. aureus and E. faecalis was different so that the antimicrobial effects of C2 in comparison with vancomycin was higher on E. faecalis but did not have statistically significant effect on S. aureus.
The results of changes in gene expression showed that the expression of DNA gyrase gene was reduced by the compound of C1 in both tested bacteria, but it was not significant in them (p=0.06). Compound C2 reduced the gene expression in each of tested bacteria and in the case of E. faecalis, reduced gene expression was significant (p<0.01).
Any brief changes in the structure of the fluoroquinolones are responsible for the development of various physiological properties, antibacterial strength and spectrum of these drugs. The synthesis and application of organofluorine (i.e., current fluoroquinolones) compounds are an important part of medical and therapeutic research.
The antibiotics have been associated globally with complications such as antibiotic resistance. The phenomenon of antibiotic resistance has led scientists to develop new types of antibiotics, although, limiting the use of these substances can delay the antibiotic resistance, the occurrence of this event is inevitable, and due to the development and growth of resistant strains, some of the antibiotics lost their effectiveness [16]. In addition, antibiotics are sometimes associated with adverse-effects, such as immuno-suppression and allergic reactions [17]. Hence, it is always necessary to produce a series of new antibiotics that bacteria are sensitive to, for the treatment of infectious diseases [18]. Two in-vitro studies; Alt S et al., and Linder KE et al., showed that S. aureus mutants resist against fluoroquinolones, which can contribute to the importance of producing new antibacterial compounds derived from fluoroquinolones to overcome the resistance of bacteria [19,20].
Lafaurie M et al., in a 10 years study showed that in the past year’s, quinolones lost their antibacterial properties on S. aureus and P. aeruginosa, two known bacteria in human infections. However, since the use of these antibiotics has been limited, this resistance has been decreasing [21]. Holla BS et al., produced several new derivatives of fluoroquinolones in India and investigated their antimicrobial effects. The efficacy of quinoline-4-carboxylic acid and its triazolothiadiazole derivatives on S. aureus and Bacillus subtilis were evaluated using a serial dilution method. In this study, the antibacterial effects of synthesised nitrofurazone (Furacin) were compared to common antibiotics. The results revealed the comparatively strong antimicrobial effects of a number of triazolothiadiazole derivatives [22]. These findings were consistent with the results of the present study, with the exception that the acetimidoyl chloride-based quinolone derivatives were assessed in the present study.
Mohammadhosseini N et al., synthesised a novel compound of 7-piperazine quinolone (1-cyclopropyl-6-fluoro-1,4-dihydro-8-methoxy-7-(2-(2-thienyl)-2-hexoxyiminoethyl)-3-methylpiperazine-1-yl)-4-oxo-3-quinolone carboxylic acid) and evaluated its effect on a number of gram-positive and gram-negative bacteria and reported that the antibacterial effects of this compound are higher even than Gatifloxacin, that is one of the best fluoroquinolones to eliminate bacterial infection [23].
In another study, Hu Y et al., examined the effect of a new type of antibiotic derived from quinolones called HT61 with a molecular weight of 400 D on a number of bacteria such as methicillin-resistant and methicillin-sensitive bacteria, including Methicillin-Resistant S. aureus (MRSA). The mechanism of this antibiotic was the depolarization of the cell membrane and the destruction of the cell wall. The results showed that this antibiotic could eliminate the mentioned bacteria six times more than conventional antibiotics. No resistant cases were observed during 50 cultivations [24].
Darehkordi A et al., after synthesis of N-aryl-2, 2, 2-trifluoroacetimidoyl piperazinyl quinolone derivatives showed that some of the new compounds have an antibacterial effect on tested gram-negative and gram-positive bacteria. In the previous study, the present authors didn’t perform MIC and DNA gyrase gene expression test properly, but at the presented study, authors find that the compounds titled C1/C2 act considerable at the expression level of a related gene [11].
Hooper DC et al., during an investigation presented that due to the emergence of drug-resistant bacteria, many studies have been carried out to synthesise new quinolones with strong effects and fewer complications. According to this principle, and to increase their effect on gram-positive bacteria, their place of action in bacteria has been attempted to be shifted from topoisomerase IV to DNA gyrase. These compounds have a relatively large group at position C7 on the piperazine ring. It has been observed that the addition of large substitutions to quinolones increases lipophilicity resulting enhancement of tissue infiltration as well as penetration into the organism, and if this group is at position C7, they increase the antibacterial effect against gram-positive bacteria [25].
Morrow BJ et al., showed that fluoroquinolones derivatives of a wide spectrum of fluorinated 4-quinolones, [7-[(3E)-3-(2-amino-1-fluoroethylidene)-1-piperidinyl]-1-cyclopropyl-6-fluoro-1,4-dihydro-8-methoxy-4-oxo-3-quinolinecarboxylic acid (JNJ-Q2) have an antibacterial effects. The antibacterial activity of this new derivative was significantly higher than that of moxifloxacin on gram-positive bacteria [26].
As Gorla MC et al., and Jenkins SG et al., showed the fluorinated quinolone has been used to treat acute skin infections and community-acquired pneumonia. This antibiotic was used according to the M32 guidelines of Clinical and Laboratory Standards Institute (CLSI), BMD and disc diffusion methods on seven important pathogenic bacteria to obtain the sensitivity or resistance of bacteria. The results indicated positive antibacterial effects of these quinolones [27,28].
Overall, the results of the above studies suggest the importance of synthesising new compounds including fluoroquinolones derivatives, to provide an appropriate strategy to counteracting antibiotic resistance in the future.
A major challenge in the synthesis of quinolone against bacteria is that the drug should have a strong effect on bacterial enzymes and not on human enzymes. For example, Aldred, KJ et al., in a study synthesised the compounds that caused the greatest effect on topoisomerase IV of Bacillus anthracis, which had a poor effect on the topoisomerase 2-alpha (DNA gyrase, subunit A), and this selectivity could have many problems with the introduction and use of antibiotics [29]. Perhaps one of the reasons that an antibiotic can affect a bacterium but not the other bacterium with the same gram staining nature is the fact that the drug (i.e., C1 in the present study) may act strongly on S. aureus but poorly on E. faecalis due to different affinity to target enzyme, etc.
As mentioned earlier, one of the problems in the treatment of bacterial infections is bacterial resistance to antibiotics, including quinolones. Recently, in the study of Wang KM et al., the isomers of Gatifloxacin, called [1-cyclopropyl-6-fluoro-1,4-dihydro-8-methoxy-7-(2-methyl-1-piperazinyl)-4-oxo-3-quinoline carboxylic acid], and a number of its derivatives were re-designed and synthesised and their antimicrobial activity was tested on strains of S.epidermidis and Klebsiella pneumoniae, which produced an outstanding antimicrobial effect [30]. Darehkordi A et al., also revealed that a new group of piperazinyl quinolone derivatives was tested against S. aureus and E. faecalis bacteria (both gram-positive) and E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Proteus mirabilis (all gram-negative) bacteria act acceptable on some bacteria. Among the various synthesised compounds, the findings of the study showed that some of these compounds had severe antimicrobial effects on all tested bacteria, indicating the effectiveness of the compounds on some gram-positive and gram-negative bacteria [31]. Saeidinia A et al., during research announced, fluoroquinolones are widely prescribed in the world for the treatment of gram-positive and gram-negative bacterial infections, and there is a rising resistance of bacteria such as E. faecalis and S. aureus to these compounds [32]. Compounds containing trifluoromethyl (phenyl) such as trifluoroacetimidoyles are of great importance and act as the building blocks for the production of some antibiotics.
In general, and according to the results of the present study and scientific reports mentioned in this context, the importance of using new compounds derived from the manipulation of antibiotics such as fluoroquinolones can lead to higher drug efficacy and, consequently, the long-term administration. Living organisms cannot simultaneously find a way to deal with different mechanisms, therefore, these compounds can be optimised in infections without worrying about drug resistance in the short-term. The difference in the effect of C2 on different genera of gram-positive bacteria and efficacy of this compound on E. faecalis compared to vancomycin, and the ineffectiveness of C2 on S. aureus versus vancomycin. The decrease in DNA gyrase gene expression and growth inhibitory effects (MIC and Disk Diffusion) of two ciprofloxacin and norfloxacin derivatives were achieved on S. aureus and E. faecalis.
Limitation
One of the limitations is the non-application of the C1/C2 compounds on the ciprofloxacin and norfloxacin-resistant bacteria and the other is the restriction of the number of bacterial strains, particularly strains isolated from infected patients. The third one was an in-vitro study in which we cannot be sure whether synthesised compounds will work in clinical cases (In-vivo) or not.
Conclusion
The results showed that the C2 compared with vancomycin and the C1 has a better antibacterial activity on E. faecalis, which is probably due to its mechanism of inhibiting DNA gyrase in this bacterium. The synthesised compounds in this study were somewhat acceptable against tested gram-positive bacteria. After further investigations, the compound of C2 may act as a new generation of antibiotics to control S. aureus and E. faecalis infections.
(+): No inhibition (Bacterial growth not affected); (-): Inhibition (Bacterial growth affected)