Introduction
There have been great advances related to impact of microbiota on cancer immunology [1]. It is a well-known fact that cancer grows in close proximity to microbiota starting from primary lesion to metastasis to distant sites [2]. It has been postulated that composition of gut microflora influences anticancer immune surveillance by both cytoprotection and immunosuppression. In addition, the gut inhabitants also play a cardinal role in determining the therapeutic response of the anticancer treatment of the patient [3]. In a nutshell, microbiota research has revolutionised both the scientific and clinical aspect of cancer immunology unveiling fascinating insights into cancer therapy.
CRC is the fourth most common cause of cancer and second most common cause of mortality among cancer worldwide. The 5 year prevalence rate of CRC is about 4.7 million cases with 8.8 lakh deaths worldwide in the year 2018 [4]. Highest incidence rates are seen in Australia, New Zealand, Canada, the United States and parts of Europe whereas the lowest risk occurs in countries like China, India, parts of Africa and South America [5]. Diet and lifestyle are crucial factors that affect the pathogenesis of CRC [6]. As inflammation plays an important role in the aetiopathogenesis of CRC, in this review we will discuss the role of gut inhabitants in carcinogenesis with main emphasis on microbial metabolism and inflammation.
The collection of microorganisms that live in peaceful co-existence with their hosts are termed as the microbiota or microflora. The microbiota colonise all surfaces of the human body that are exposed to external environment including skin, genitourinary, gastrointestinal, and respiratory tracts [7]. The large intestine contains the densest microbial community containing over 70% of all the microbes in the human body [8]. Anerobic bacteria, the Bacteroidetes and the Firmicutes dominate the large colon, whereas Proteobacteria, Verrucomicrobia and Actinobacteria are present in minor proportions [9]. The gut microbes which appear at birth are similar to mother’s vaginal microflora. The microflora undergo changes and start resembling the adult’s flora after 1 year of age [10]. There are several factors like genetic makeup and dietary habits of the individual which influence the composition of gut flora [11,12]. There are two types of fecal microflora in the general population namely Low Gene Count (LGC) and High Gene Count (HGC). The LGC microflora shows less diversity and has more of bacteroides and less of Firmicutes [13]. On the other hand HGC are characterised by more diversity of gut flora and more of Firmicutes. In 2013, Cotillard A et al., postulated that individuals with obesity have LGC microbe community and weight loss diet improved microbial diversity, and which ultimately drifted towards HGC [14]. This result emphasises on the importance of diet on modulating the composition of gut flora.
Importance of Microbiota in The Intestine
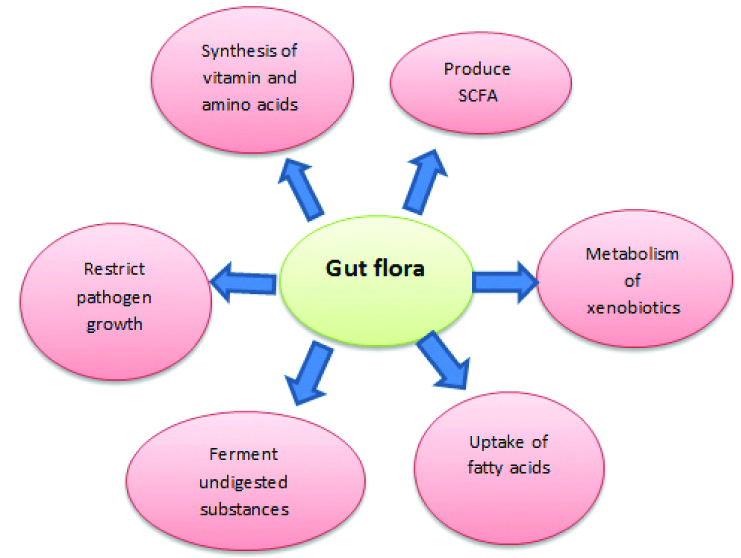
[Table/Fig-1] depicts the importance of microbiota in the intestine. The microbiota benefits the host’s gut by restricting the pathogen’s growth by colonising and occupying the attachment sites in the gut, and consuming the available nutrients [15]. It not only produces but also stimulates the host to produce antimicrobial substances (AMP-Antimicrobial peptides). These AMPs prevent overgrowth of commensals and prevent invasion from pathogens [16]. The gram positive bacteria such as Lactobacillus prevent Listeria infection in vitro through production of antimicrobial substances and modulation of immune responses of the epithelial cells [17]. The microbes help in maximising the caloric availability of ingested nutrients by extracting additional calories [18]. They help in fermenting the undigested dietary components (mucin, non-starch polysaccharides and resistant starch) into short chain fatty acids like acetate, propionate and butyrate in the ratio 3:1:1 [19]. In addition, the microbes also promote the uptake of fatty acids into adipocytes by suppressing the inhibition of lipoprotein lipase [20]. The metabolism of xenobiotics including pharmaceuticals is associated with contribution from both host and microbiota. Therefore, a new concept of pharmacometabonomics has now emerged. The gut inhabitants are associated with degradation of dietary oxalates that underlines their role in prophylactic treatment in calcium oxalate nephrolithiasis [21,22]. It also degrades hydrazine, a known industrial toxin and acts as a detoxifier [23].
Importance of microbiota.
SCFA: Short chain fatty acid
Role of Microbiota in Aetiopathogenesis of CRC
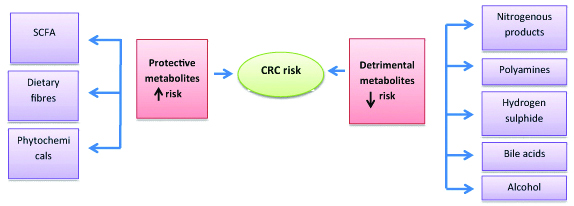
[Table/Fig-2] shows the role of microbial metabolism in the aetiopathogenesis of CRC. There are several models for microbial involvement in colorectal carcinogenesis. The first model states that single microbe causes disease aetiology, for example Helicobacter pylori causing gastritis. The second model says that initially host factors initiate disease pathology followed by change and alteration in microbiota leading to dysbiosis. The third model suggests that some gut inhabitants can aggravate or reduce the intensity of certain infections however clinical relevance remains debatable [24]. Accumulating evidence suggests that microbial metabolism influences CRC aetiopathogenesis [24]. These metabolites produced by gut flora impact carcinogenesis by various mechanisms.
Protective Metabolites
1. Short Chain Fatty Acids (SCFAs)
The major bacterial fermentation products are the SCFAs namely butyrate, propionate and acetate. Butyrate is used as energy source by gut epithelium [25]. Butyrate and propionate when transported into host cells participate in epigenetic modification by directly inhibiting histone deacetylases in colonocytes and immune cells [26]. This leads to hyperacetylation of histones which further results in down regulation of pro-inflammatory cytokines (IL-6 and IL-12) in colonic macrophages which accounts for their anti-inflammatory effect [27]. Additionally, histone H3 acetylation in the promoter and enhancer region of Forkhead box P3 (FOXP3), leads to increased expression of FOXP3 and hence differentiation of regulatory T cells [28]. These SCFAs can also exert indirect effects by stimulating G protein-coupled receptors (GPCRs) on the surface of colonocytes and immune cells [28]. Hence, both Histone deacetylases (HDAC) inhibition and GPCR signaling results in an increase in total colonic regulatory T cell (cTReg) number and the production of anti-inflammatory Interleukin-10 (IL-10) and Transforming Growth Factor-β (TGFβ). In addition, HDAC inhibition is also thought to promote apoptosis of CRC cells [29]. Hence, butyrate is shown to prevent carcinogenesis by its anti-inflammatory and anti-apoptotic effect. Butyrate also assists the assembly of tight junctions by the activation of AMPK and thereby promotes the intestinal barrier function [30]. SCFAs lower the pH of colonic lumen resulting in various effects. This lower pH decreases the carcinogenic activity of secondary bile acids and the bacterial enzyme 7-α-dehydroxylase which converts primary to secondary bile acids. Since the solubility of calcium also increases at lower pH, higher concentration is available for binding with bile acids. As a result it has been seen that lower fecal pH is associated with lower risk of colon cancer [31].
2. Dietary Fibers
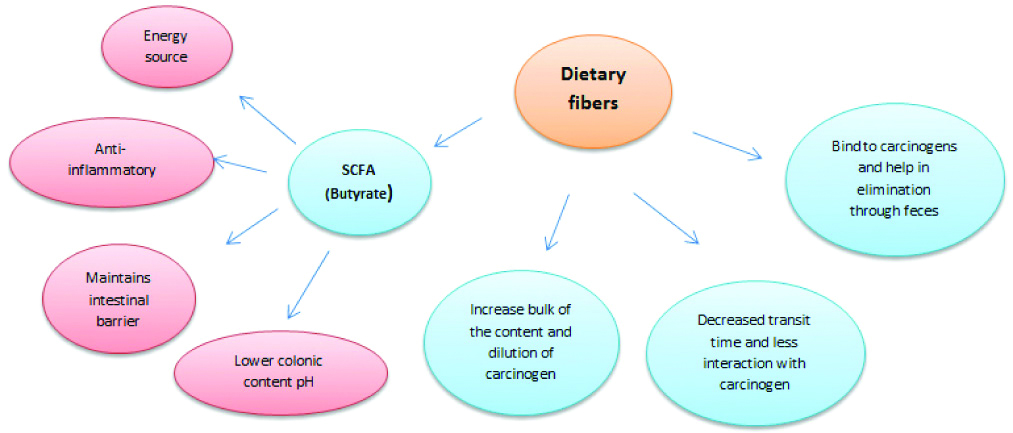
[Table/Fig-3] shows the role of dietary fibers in protection against CRC. The bacterial anaerobic fermentation of dietary fibers converts them into SCFAs, mainly acetate, propionate, butyrate and gases like hydrogen, carbon dioxide, methane, and hydrogen sulphide [32]. Most fermentation occurs in the proximal large bowel and the luminal environment where the pH turns acidic due to increased formation of SCFAs [33]. Apart from indirect benefit of generating SCFA, dietary fibers have many direct advantages. The undigested fibers bind to carcinogens and hence are eliminated through feces [34]. Dietary fibers increase the bulk of feces and results in shorter transit times with minimal interaction between carcinogens and mucosal cells [35]. As the bulk of the colon content increases, it also results in dilution of carcinogens which serves as another added benefit [36].
Role of microbial metabolism in the aetiopathogeneis of colorectal cancer.
SCFA: Short chain fatty acid; CRC: Colorectal cancer
Role of dietary fibers in protection against colorectal cancer.
3. Phytochemicals
Present as glycosides in the large intestine where it is transformed by gut microbiota. These newly formed metabolites inhibit pro-inflammatory mediators like TNF-α, NF-κβ and prostanoids and thereby contribute to healthy gut environment [37].
Detrimental Metabolites
1. Nitrogenous Products
In high protein diets, nitrogenous products produced by fermentation of protein in colon can cause deleterious effects in the gut environment [38]. These compounds are formed endogenously via acid-driven nitrosation of amines in the stomach and microbial fermentation in the intestine [39]. It is suggested that the nitro reductases and nitrate reductases that catalyse nitrosation reactions are encoded by proteobacteria. The nitrogenous products which can be formed are branched-chain fatty acids, phenylacetic acid and N-nitroso compounds (NOCs) [40]. NOCs exert carcinogenic effects via DNA alkylation that causes mutations and leads to cancer [41].
2. Polyamines
Polyamines (putrescine, spermidine and spermine) synthesised by the gut bacteria and the host cells, are involved in the maintenance of the structural integrity of membranes and nucleic acids, gene regulation and translation [42,43]. Certain gut bacteria (such as enterotoxigenic Bacteroides fragilis) also upregulate polyamine production by host cells thus leading to high levels of polyamines [44]. The polyamine catabolism can lead to oxidative stress which is further associated development of cancer.
3. Hydrogen Sulphide
Studies suggest that by-products of metabolism of dietary proteins (suphur containing amino acids and taurine) such as hydrogen sulphide produced by sulfate reducing bacteria can be carcinogenic to the gut mucosa due to its cytotoxic effects [45,46]. Moreover, it has been reported that sulphide also prevents oxidation of butyrate, and is genotoxic due to ROS production [47,48]. Any modification of microbiota towards non-sulphate reducing bacteria (methanogenic bacteria) reduces the risk for colon carcinoma.
4. Bile Acids
High-fat diet lead to an increase in bile secretion, and increased fecal bile acid concentrations and are associated with the higher incidence of CRC [49]. When diet supplemented with deoxycholic acid is fed to rats, a decrease in the production of SCFAs along with major changes in the composition of the gut microflora was observed [50].
The primary bile acids cholic acid and chenodeoxycholic acid are produced in the liver from cholesterol, are conjugated to glycine or taurine (which render the bile acids more hydrophilic) and are excreted into the duodenum to facilitate fat digestion. Majority of the primary bile acids are reabsorbed in the terminal ileum for enterohepatic circulation. However, the fraction of bile acid (approximately 5% of the total pool) which skips enterohepatic circulation undergoes extensive transformation by the microbiota in the large intestine [51]. Bacterial strains and the methanogenic archaea have bile salt hydrolases which cleave glycine and taurine residues from the primary bile acids along with dehydrogenation and dehydroxylations converting them into secondary bile acids (deoxycholic acid and lithocholic acid) [52].
Bile acids have strong antimicrobial activities due to their amphipathic properties. They can damage bacterial cell membranes and are likely to modify the composition of the gut microbiota. Owing to the production of Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS) that causes DNA damage, bile acids have been implicated in carcinogenesis of the intestinal tract [53]. Secondary bile acids can promote carcinogenesis by various mechanisms. They are more hydrophobic and thus more potent at disrupting cell membranes. This leads to the generation of ROS via the activation of membrane-associated proteins such as NADPH oxidases and phospholipase A2 [54]. They can also induce NF-kB activation in intestinal epithelial cells and trigger proinflammatory cascade [55]. In-vitro studies indicate that their long term exposure can induce carcinogenesis by decreasing apoptosis and enhancing epithelial cell proliferation [49]. Thus, it can be postulated that both primary and secondary bile acids have potential as therapeutic targets for alleviating inflammation which promotes carcinogenesis.
Various other factors have also been implicated in linking high fat diet to CRC. In 2012, Devkota S et al., reported that taurine conjugation of primary bile acids increases due to high fat diet [56]. This increases the growth of sulphur reducing bacterium Bilophila wadsworthia that further promotes inflammation [57]. Furthermore, it has been observed that Clostridium, Ruminococcus can convert chenodeoxycholic acid (primary bile acid) into ursodeoxycholic acid which can have beneficial effects in humans [58]. However, in the presence of 7b-dehydroxylase enzyme, the given strain can also convert ursodeoxycholic acid to the potentially toxic lithocholic acid. Further studies are required to validate microbe-based bile acid metabolism approach for CRC prophylaxis, and identifying the bacterium which can form ursodeoxycholic acid.
5. Ethanol (Alcohol)
Excessive consumption of ethanol is considered to be an important risk factor for several cancers [59]. Microbial metabolism may contribute to its toxicity, especially in the upper gastrointestinal tract. Acetaldehyde, a product of ethanol oxidation produced by microbial metabolism can cause DNA damage and degradation of the vitamin folate and is highly toxic and carcinogenic [60,61].
Microbiota and Inflammation
[Table/Fig-4] shows the determinants of inflammation and their role in colorectal carcinogenesis. The colonic mucosa is constantly exposed to the gut microbiota and its metabolites with potential to cause chronic low-grade inflammation [62]. Therefore, the role of microbiota in aetiopathogenesis of CRC becomes important as chronic inflammation is a risk factor for CRC [63]. It is seen that germ-free ApcMin (mice having mutation in one copy of the tumour suppressor gene Apc) exhibit a two-fold reduction in small intestinal adenomas compared with ApcMin mice with normal microbiota composition [64]. This highlights the role of microbiota in triggering inflammation.
Determinants of inflammation and its role in colorectal carcinogenesis.
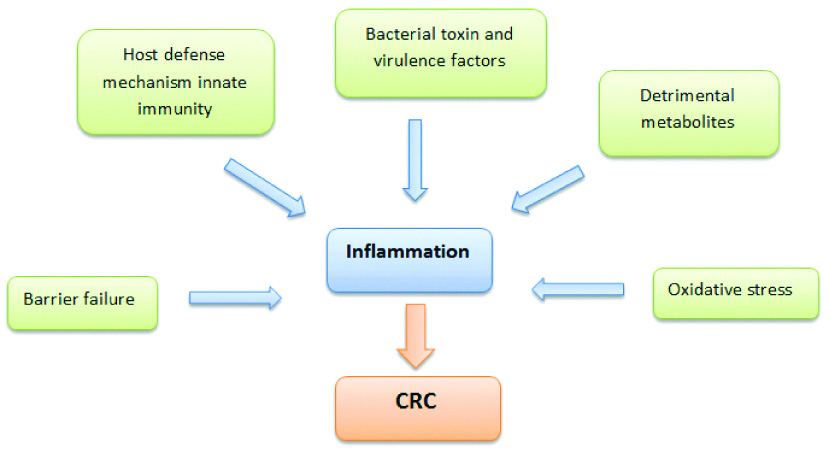
The role of microbiota in inflammation is also underscored by the fact that altered Toll-Like Receptor-4 (TLR4) signaling is associated with CRC progression [65]. TLR4 is type of PRRs (Pattern Recognition Receptors) present in host that recognises Microorganism-Associated Molecular Patterns (MAMPs) such as Lipopolysaccharides (LPS), nucleic acids elicits an inflammatory response [66].
One of the most pertinent mechanisms for microbial tumourigenesis is loss of barrier function in the colonic epithelium leading to enhanced microbiota-host interactions. The initial activation of β-catenin and mutation of the APC gene in colorectal tumourigenesis may result in a loss of barrier function in the colonic epithelium leading to translocation of microbial products into the tumour microenvironment and colonisation of invasive-adherent bacteria at neoplastic sites [67]. Barrier failure can result from primary defects in genes that encode proteins that are essential to maintain a functional barrier, or from secondary defects owing to infection, inflammation and carcinogenesis. Ulcerative colitis is also associated with defects in intestinal barrier which predisposes the affected individuals to increased risk of cancer [68]. This underlines the importance of barrier permeability in carcinogenesis and inflammation.
Fusobacterium nucleatum
High incidence of gram negative anerobe in adenoma implicates its role in tumour growth initiation and aetiopathogenesis of CRC [69]. Studies suggest that its role in commencing carcinogenesis can be attributed to binding of its FadA antigen with the E-cadherin on colonocytes leading to activation of Wnt/β-catenin pathway [70,71]. This leads to uncontrolled cell division, loss of cell polarity and MSI tumour phenotype. It has also be known to bestow chemotherapeutic resistance by stimulating TLR-4 signaling and triggering autophagy by modulating regulatory microRNA [72]. Fusobacterium nucleatum is the most widespread strain associated with CRC and is associated with initiation, severity and modulating chemotherapeutic responses of the tumour [72].
Escherichia coli
E. coli is the most common cultivable, gram-negative, aero-anaerobic commensal gut bacteria. It has four different phylogroups depending on the presence of virulence factors. Out of them B2 and D are pathogenic and are involved in various diseases [73]. These strains synthesise several toxins called cyclomodulins such as Cytolethal Distending Toxins (CDT), Cytotoxic Necrotising Factor (CNF), cycle inhibiting factor, and colibactin [74]. Cyclomodulins are genotoxic and/or modulate cell-cycle progression, proliferation, cell differentiation, and apoptosis [75]. Colibactin, a hybrid polyketide nonribosomal peptide encoded by the pks genomic island can induce DNA double-strand breaks in the host cell and thereby activate DNA damage signalling cascades [76]. This can further lead to chronic mitotic and chromosomal aberrations as well as an increased frequency of gene mutation [77].
Enterotoxigenic Bacteroides fragilis
Enterotoxigenic Bacteroides fragilis is also implicated in intestinal carcinogenesis [78]. It is anerobic bacteria constituting 1% of gut microbiota [79]. It has two subtypes nontoxigenic and enterotoxigenic. The enterotoxigenic type is associated with production of BFT toxin that induces Spermine Oxidase (SPO) generating ROS which leads to DNA damage in intestinal epithelial cells [80,81].
Recent Updates
Li Shizhen gave the notion of using yellow soup (containing fecal matter) to treat abdominal diseases originated in China. Currently, Fecal Microbiota Transplantation (FMT) has become recognised and has shown promising results for treating patients with recurrent Clostridium difficile infection [82]. Few studies on patients with ulcerative colitis, have shown promising results, but its efficacy for treatment for Inflammatory Bowel Disease (IBD) remains unclear [83,84]. Although various studies have been conducted on investigating the role of FMT in patients with IBS, Hepatic Encephalopathy (HE), Metabolic syndrome, Autism, Graft Versus Host Disease (GVHD), no conclusive results have been obtained [85-88]. In 2017, Zhou YJ et al., showed that intestinal microflora belonging to phyla such as Actinobacteria, Proteobacteria and Firmicutes have anticarcinogenic effect against solid tumours and leukaemia [89]. In 2016, Hu Y et al., illustrated the role of resistant starch as protective agent against colitis associated with CRC in rodent model [90].
Conclusion
The role of the microbiota in CRC has become quite blatant and thereby delineates a new approach aimed at ameliorating the therapeutic management of patients with CRC. It is seen that the advancement to CRC is influenced by the metabolic output of the microbiota. These protective and detrimental metabolites can significantly influence gut microenvironment through the specific interactions at the mucosal level. Therefore, the diverse and healthy microbiota is essential in establishing stable and healthy gut microbiome and preventing dysbiosis characterised by a reduction in microbial diversity. The dietary manipulation has the ability to modify microbiome composition and offer desired benefits. Therefore, there is a need for identifying, analysing and assessing such strains for their long term effect on the health of the individual. There are many challenges which need to be addressed like the diagnostic efficacy of fecal microbiome is questionable as it mirrors the intraluminal microbiome and not mucosal microbiome. Moreover there are differences in the results of the numerous studies analysing the gut microbiome making standardisation of microbiota signature associated with colorectal carcinoma even more difficult. In addition to this, greater than 80% of our gut inhabitants are not cultivable. It thus becomes important to explore new molecular methods to characterise the microbial ecology, physiology and functional capacity.
[1]. Kroemer G, Zitvogel L, Cancer immunotherapy in 2017: The breakthrough of the microbiotaNat Rev Immunol 2018 18(2):87-88.10.1038/nri.2018.429379189 [Google Scholar] [CrossRef] [PubMed]
[2]. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Analysis of Fusobacterium persistence and antibiotic response in colorectal cancerScience 2017 358(6369):1443-48.10.1126/science.aal524029170280 [Google Scholar] [CrossRef] [PubMed]
[3]. Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, Intestinal microbiota and relapse after hematopoietic-cell transplantationJ Clin Oncol Off J Am Soc Clin Oncol 2017 35(15):1650-59.10.1200/JCO.2016.70.334828296584 [Google Scholar] [CrossRef] [PubMed]
[4]. Cancer today [Internet]. 2019 [cited 2019 Jan 30]. Available from: http://gco.iarc.fr/today/home [Google Scholar]
[5]. Haggar F, Boushey R, Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factorsClin Colon Rectal Surg 2009 22(04):191-97.10.1055/s-0029-124245821037809 [Google Scholar] [CrossRef] [PubMed]
[6]. Wang ZH, Fang JY, Colorectal cancer in inflammatory bowel disease: Epidemiology, pathogenesis and surveillanceGastrointest Tumours 2014 1(3):146-54.10.1159/00036530926674110 [Google Scholar] [CrossRef] [PubMed]
[7]. Kunz C, Kuntz S, Rudloff S, Intestinal Flora. In: Goldberg G, Prentice A, Prentice A, Filteau S, Simondon K, editorsBreast-Feeding: Early Influences on Later Health [Internet] 2009 [cited 2019 Apr 11] DordrechtSpringer Netherlands:67-79.Available from: http://link.springer.com/10.1007/978-1-4020-8749-3_610.1007/978-1-4020-8749-3_619227535 [Google Scholar] [CrossRef] [PubMed]
[8]. Hillman ET, Lu H, Yao T, Nakatsu CH, Microbial ecology along the gastrointestinal tractMicrobes Environ 2017 32(4):300-13.10.1264/jsme2.ME1701729129876 [Google Scholar] [CrossRef] [PubMed]
[9]. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Diversity of the human intestinal microbial floraScience 2005 308(5728):1635-38.10.1126/science.111059115831718 [Google Scholar] [CrossRef] [PubMed]
[10]. Mackie RI, Sghir A, Gaskins HR, Developmental microbial ecology of the neonatal gastrointestinal tractAm J Clin Nutr 1999 69(5):1035s-45s.10.1093/ajcn/69.5.1035s10232646 [Google Scholar] [CrossRef] [PubMed]
[11]. Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, Influence of diet on the gut microbiome and implications for human healthJ Transl Med 2017 15(1):7310.1186/s12967-017-1175-y28388917 [Google Scholar] [CrossRef] [PubMed]
[12]. Das B, Ghosh TS, Kedia S, Rampal R, Saxena S, Bag S, Analysis of the gut microbiome of rural and urban healthy indians living in sea level and high altitude areasSci Rep 2018 8(1):1010410.1038/s41598-018-28550-329973712 [Google Scholar] [CrossRef] [PubMed]
[13]. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural AfricaProc Natl Acad Sci USA 2010 107(33):14691-96.10.1073/pnas.100596310720679230 [Google Scholar] [CrossRef] [PubMed]
[14]. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Dietary intervention impact on gut microbial gene richnessNature 2013 500(7464):585-88.10.1038/nature1248023985875 [Google Scholar] [CrossRef] [PubMed]
[15]. Kho ZY, Lal SK, The human gut microbiome- A potential controller of wellness and diseaseFront Microbiol 2018 9:183510.3389/fmicb.2018.0183530154767 [Google Scholar] [CrossRef] [PubMed]
[16]. Thursby E, Juge N, Introduction to the human gut microbiotaBiochem J 2017 474(11):1823-36.10.1042/BCJ2016051028512250 [Google Scholar] [CrossRef] [PubMed]
[17]. Corr SC, Gahan CGM, Hill C, Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune responseFEMS Immunol Med Microbiol 2007 50(3):380-88.10.1111/j.1574-695X.2007.00264.x17537177 [Google Scholar] [CrossRef] [PubMed]
[18]. Flint HJ, Scott KP, Louis P, Duncan SH, The role of the gut microbiota in nutrition and healthNat Rev Gastroenterol Hepatol 2012 9(10):577-89.10.1038/nrgastro.2012.15622945443 [Google Scholar] [CrossRef] [PubMed]
[19]. Canfora EE, Jocken JW, Blaak EE, Short-chain fatty acids in control of body weight and insulin sensitivityNat Rev Endocrinol 2015 11(10):577-91.10.1038/nrendo.2015.12826260141 [Google Scholar] [CrossRef] [PubMed]
[20]. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, The gut microbiota as an environmental factor that regulates fat storageProc Natl Acad Sci USA 2004 101(44):15718-23.10.1073/pnas.040707610115505215 [Google Scholar] [CrossRef] [PubMed]
[21]. Sidhu H, Allison MJ, Chow JM, Clark A, Peck AB, Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenesJ Urol 2001 166(4):1487-91.10.1016/S0022-5347(05)65817-X [Google Scholar] [CrossRef]
[22]. Hoppe B, Beck B, Gatter N, von Unruh G, Tischer A, Hesse A, Oxalobacterformigenes: A potential tool for the treatment of primary hyperoxaluria type 1Kidney Int 2006 70(7):1305-11.10.1038/sj.ki.500170716850020 [Google Scholar] [CrossRef] [PubMed]
[23]. Swann J, Wang Y, Abecia L, Costabile A, Tuohy K, Gibson G, Gut microbiome modulates the toxicity of hydrazine: A metabonomic studyMol Biosyst 2009 5(4):351-55.10.1039/b811468d19396371 [Google Scholar] [CrossRef] [PubMed]
[24]. Sears CL, Garrett WS, Microbes, microbiota, and colon cancerCell Host Microbe 2014 15(3):317-28.10.1016/j.chom.2014.02.00724629338 [Google Scholar] [CrossRef] [PubMed]
[25]. Hague A, Butt AJ, Paraskeva C, The role of butyrate in human colonic epithelial cells: An energy source or inducer of differentiation and apoptosis?Proc Nutr Soc 1996 55(3):937-43.10.1079/PNS199600909004335 [Google Scholar] [CrossRef] [PubMed]
[26]. Fung KYC, Cosgrove L, Lockett T, Head R, Topping DL, A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrateBr J Nutr 2012 108(5):820-31.10.1017/S000711451200194822676885 [Google Scholar] [CrossRef] [PubMed]
[27]. Chang PV, Hao L, Offermanns S, Medzhitov R, The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibitionProc Natl Acad Sci USA 2014 111(6):2247-52.10.1073/pnas.132226911124390544 [Google Scholar] [CrossRef] [PubMed]
[28]. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesisImmunity 2014 40(1):128-39.10.1016/j.immuni.2013.12.00724412617 [Google Scholar] [CrossRef] [PubMed]
[29]. Wilson AJ, Chueh AC, Tögel L, Corner GA, Ahmed N, Goel S, Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene inductionCancer Res 2010 70(2):609-20.10.1158/0008-5472.CAN-09-232720068171 [Google Scholar] [CrossRef] [PubMed]
[30]. Peng L, Li ZR, Green RS, Holzman IR, Lin J, Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayersJ Nutr 2009 139(9):1619-25.10.3945/jn.109.10463819625695 [Google Scholar] [CrossRef] [PubMed]
[31]. Samelson SL, Nelson RL, Nyhus LM, Protective role of faecal pH in experimental colon carcinogenesisJ R Soc Med 1985 78(3):230-33.10.1177/0141076885078003113973888 [Google Scholar] [CrossRef] [PubMed]
[32]. Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Gut microbiota functions: Metabolism of nutrients and other food componentsEur J Nutr 2018 57(1):01-24.10.1007/s00394-017-1445-828393285 [Google Scholar] [CrossRef] [PubMed]
[33]. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM, The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolismJ Lipid Res 2013 54(9):2325-40.10.1194/jlr.R03601223821742 [Google Scholar] [CrossRef] [PubMed]
[34]. Jacobs LR, Relationship between dietary fiber and cancer: Metabolic, physiologic, and cellular mechanismsExp Biol Med 1986 183(3):299-310.10.3181/00379727-183-424233025886 [Google Scholar] [CrossRef] [PubMed]
[35]. Harris PJ, Ferguson LR, Dietary fibre: Its composition and role in protection against colorectal cancerMutat Res Mol Mech Mutagen 1993 290(1):97-110.10.1016/0027-5107(93)90037-G [Google Scholar] [CrossRef]
[36]. Koriech OM, Diet and cancerJ Fam Community Med 1994 1(1):02-11. [Google Scholar]
[37]. Surh YJ, Chun KS, Cha HH, Han SS, Keum YS, Park KK, Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-κB activationMutat Res Mol Mech Mutagen 2001 480-81:243-68.10.1016/S0027-5107(01)00183-X [Google Scholar] [CrossRef]
[38]. Delimaris I, Adverse Effects associated with protein intake above the recommended dietary allowance for adultsISRN Nutr 2013 2013:12692910.5402/2013/12692924967251 [Google Scholar] [CrossRef] [PubMed]
[39]. Rowland RH, Ian R, Metabolic activities of the gut microflora in relation to cancerMicrob Ecol Health Dis 2000 12(2):179-85.10.1080/089106000750060431 [Google Scholar] [CrossRef]
[40]. Windey K, De Preter V, Verbeke K, Relevance of protein fermentation to gut healthMol Nutr Food Res 2012 56(1):184-96.10.1002/mnfr.20110054222121108 [Google Scholar] [CrossRef] [PubMed]
[41]. Bartsch H, Ohshima H, Shuker DE, Pignatelli B, Calmels S, Exposure of humans to endogenous N-nitroso compounds: Implications in cancer etiologyMutat Res 1990 238(3):255-67.10.1016/0165-1110(90)90017-6 [Google Scholar] [CrossRef]
[42]. Di Martino ML, Campilongo R, Casalino M, Micheli G, Colonna B, Prosseda G, Polyamines: Emerging players in bacteria-host interactionsInt J Med Microbiol 2013 303(8):484-91.10.1016/j.ijmm.2013.06.00823871215 [Google Scholar] [CrossRef] [PubMed]
[43]. Hanfrey CC, Pearson BM, Hazeldine S, Lee J, Gaskin DJ, Woster PM, Alternative spermidine biosynthetic route is critical for growth of Campylobacter jejuni and is the dominant polyamine pathway in human gut microbiotaJ Biol Chem 2011 286(50):43301-12.10.1074/jbc.M111.30783522025614 [Google Scholar] [CrossRef] [PubMed]
[44]. Pegg AE, Toxicity of polyamines and their metabolic productsChem Res Toxicol 2013 26(12):1782-800.10.1021/tx400316s24224555 [Google Scholar] [CrossRef] [PubMed]
[45]. Magee EA, Richardson CJ, Hughes R, Cummings JH, Contribution of dietary protein to sulfide production in the large intestine: An in vitro and a controlled feeding study in humansAm J Clin Nutr 2000 72(6):1488-94.10.1093/ajcn/72.6.148811101476 [Google Scholar] [CrossRef] [PubMed]
[46]. Deplancke B, Gaskins HR, Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cellsFASEB J Off Publ Fed Am Soc Exp Biol 2003 17(10):1310-12.10.1096/fj.02-0883fje12738807 [Google Scholar] [CrossRef] [PubMed]
[47]. Roediger WE, Moore J, Babidge W, Colonic sulfide in pathogenesis and treatment of ulcerative colitisDig Dis Sci 1997 42(8):1571-79.10.1023/A:10188517239209286219 [Google Scholar] [CrossRef] [PubMed]
[48]. Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR, DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cellsEnviron Mol Mutagen 2010 51(4):304-14.10.1002/em.2054620120018 [Google Scholar] [CrossRef] [PubMed]
[49]. Barrasa JI, Olmo N, Lizarbe MA, Turnay J, Bile acids in the colon, from healthy to cytotoxic moleculesToxicol Vitro Int J Publ Assoc BIBRA 2013 27(2):964-77.10.1016/j.tiv.2012.12.02023274766 [Google Scholar] [CrossRef] [PubMed]
[50]. Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Bile acid is a host factor that regulates the composition of the cecal microbiota in ratsGastroenterology 2011 141(5):1773-81.10.1053/j.gastro.2011.07.04621839040 [Google Scholar] [CrossRef] [PubMed]
[51]. Ridlon JM, Kang DJ, Hylemon PB, Bile salt biotransformations by human intestinal bacteriaJ Lipid Res 2006 47(2):241-59.10.1194/jlr.R500013-JLR20016299351 [Google Scholar] [CrossRef] [PubMed]
[52]. Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR, Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiomeProc Natl Acad Sci USA 2008 105(36):13580-85.10.1073/pnas.080443710518757757 [Google Scholar] [CrossRef] [PubMed]
[53]. Bernstein H, Bernstein C, Payne CM, Dvorak K, Bile acids as endogenous etiologic agents in gastrointestinal cancerWorld J Gastroenterol 2009 15(27):3329-40.10.3748/wjg.15.332919610133 [Google Scholar] [CrossRef] [PubMed]
[54]. Májer F, Sharma R, Mullins C, Keogh L, Phipps S, Duggan , New highly toxic bile acids derived from deoxycholic acid, chenodeoxycholic acid and lithocholic acidBioorg Med Chem 2014 22(1):256-68.10.1016/j.bmc.2013.11.02924332653 [Google Scholar] [CrossRef] [PubMed]
[55]. Da Silva M, Jaggers GK, Verstraeten SV, Erlejman AG, Fraga CG, Oteiza PI, Large procyanidins prevent bile-acid-induced oxidant production and membrane-initiated ERK1/2, p38, and Akt activation in Caco-2 cellsFree Radic Biol Med 2012 52(1):151-59.10.1016/j.freeradbiomed.2011.10.43622074817 [Google Scholar] [CrossRef] [PubMed]
[56]. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- miceNature 2012 487(7405):104-08.10.1038/nature1122522722865 [Google Scholar] [CrossRef] [PubMed]
[57]. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Diet rapidly and reproducibly alters the human gut microbiomeNature 2014 505(7484):559-63.10.1038/nature1282024336217 [Google Scholar] [CrossRef] [PubMed]
[58]. Lee J-Y, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A, Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colonJ Lipid Res 2013 54(11):3062-69.10.1194/jlr.M03983423729502 [Google Scholar] [CrossRef] [PubMed]
[59]. Seitz HK, Pöschl G, Simanowski UA, Alcohol and cancerRecent Dev Alcohol Off Publ Am Med Soc Alcohol Res Soc Alcohol Natl Counc Alcohol 1998 14:67-95.10.1007/0-306-47148-5_4 [Google Scholar] [CrossRef]
[60]. Salaspuro M, INVITED REVIEW Microbial metabolism of ethanol and acetaldehyde and clinical consequencesAddict Biol 1997 2(1):35-46.10.1080/1355621977284026735439 [Google Scholar] [CrossRef] [PubMed]
[61]. Homann N, Tillonen J, Salaspuro M, Microbially produced acetaldehyde from ethanol may increase the risk of colon cancer via folate deficiencyInt J Cancer 2000 86(2):169-73.10.1002/(SICI)1097-0215(20000415)86:2<169::AID-IJC4>3.0.CO;2-3 [Google Scholar] [CrossRef]
[62]. Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeabilityGut 2009 58(8):1091-103.10.1136/gut.2008.16588619240062 [Google Scholar] [CrossRef] [PubMed]
[63]. Lin C, Cai X, Zhang J, Wang W, Sheng Q, Hua H, Role of gut microbiota in the development and treatment of colorectal cancerDigestion 2019 100(1):72-78.10.1159/00049405230332668 [Google Scholar] [CrossRef] [PubMed]
[64]. Dove WF, Clipson L, Gould KA, Luongo C, Marshall DJ, Moser AR, Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) statusCancer Res 1997 57(5):812-14. [Google Scholar]
[65]. Neal MD, Sodhi CP, Jia H, Dyer M, Egan CE, Yazji I, Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosisJ Biol Chem 2012 287(44):37296-308.10.1074/jbc.M112.37588122955282 [Google Scholar] [CrossRef] [PubMed]
[66]. Schwabe RF, Jobin C, The microbiome and cancerNat Rev Cancer 2013 13(11):800-12.10.1038/nrc361024132111 [Google Scholar] [CrossRef] [PubMed]
[67]. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growthNature 2012 491(7423):254-58.10.1038/nature1146523034650 [Google Scholar] [CrossRef] [PubMed]
[68]. Khor B, Gardet A, Xavier RJ, Genetics and pathogenesis of inflammatory bowel diseaseNature 2011 474(7351):307-17.10.1038/nature1020921677747 [Google Scholar] [CrossRef] [PubMed]
[69]. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Genomic analysis identifies association of Fusobacterium with colorectal carcinomaGenome Res 2012 22(2):292-98.10.1101/gr.126573.11122009990 [Google Scholar] [CrossRef] [PubMed]
[70]. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Fusobacterium nucleatum potentiates intestinal tumourigenesis and modulates the tumour-immune microenvironmentCell Host Microbe 2013 14(2):207-15.10.1016/j.chom.2013.07.00723954159 [Google Scholar] [CrossRef] [PubMed]
[71]. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW, Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesinCell Host Microbe 2013 14(2):195-206.10.1016/j.chom.2013.07.01223954158 [Google Scholar] [CrossRef] [PubMed]
[72]. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagyCell 2017 170(3):548-63.e16.10.1016/j.cell.2017.07.00828753429 [Google Scholar] [CrossRef] [PubMed]
[73]. Chakraborty A, Saralaya V, Adhikari P, Shenoy S, Baliga S, Hegde A, Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in south indian populationAnn Med Health Sci Res 2015 5(4):241-46.10.4103/2141-9248.16019226229711 [Google Scholar] [CrossRef] [PubMed]
[74]. Dubois D, Delmas J, Cady A, Robin F, Sivignon A, Oswald E, Cyclomodulins in urosepsis strains of Escherichia coliJ Clin Microbiol 2010 48(6):2122-29.10.1128/JCM.02365-0920375237 [Google Scholar] [CrossRef] [PubMed]
[75]. El-Aouar Filho RA, Nicolas A, De Paula Castro TL, Deplanche M, De Carvalho Azevedo VA, Goossens PL, Heterogeneous family of cyclomodulins: smart weapons that allow bacteria to hijack the eukaryotic cell cycle and promote infectionsFront Cell Infect Microbiol 2017 7:20810.3389/fcimb.2017.0020828589102 [Google Scholar] [CrossRef] [PubMed]
[76]. Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Intestinal inflammation targets cancer-inducing activity of the microbiotaScience 2012 338(6103):120-23.10.1126/science.122482022903521 [Google Scholar] [CrossRef] [PubMed]
[77]. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP, Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cellsProc Natl Acad Sci USA 2010 107(25):11537-42.10.1073/pnas.100126110720534522 [Google Scholar] [CrossRef] [PubMed]
[78]. Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Microbial dysbiosis in colorectal cancer (CRC) patientsPloS One 2011 6(1):e1639310.1371/journal.pone.001639321297998 [Google Scholar] [CrossRef] [PubMed]
[79]. Huang JY, Lee SM, Mazmanian SK, The human commensal Bacteroides fragilis binds intestinal mucinAnaerobe 2011 17(4):137-41.10.1016/j.anaerobe.2011.05.01721664470 [Google Scholar] [CrossRef] [PubMed]
[80]. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, The bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patientsClin Infect Dis 2015 60(2):208-15.10.1093/cid/ciu78725305284 [Google Scholar] [CrossRef] [PubMed]
[81]. Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumourigenesisProc Natl Acad Sci USA 2011 108(37):15354-59.10.1073/pnas.101020310821876161 [Google Scholar] [CrossRef] [PubMed]
[82]. Debast SB, Bauer MP, Kuijper EJ, Committee. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infectionClinical Microbiology and Infection 2014 20:01-26.10.1111/1469-0691.1241824118601 [Google Scholar] [CrossRef] [PubMed]
[83]. Costello SP, Soo W, Bryant RV, Jairath V, Hart AL, Andrews JM, Systematic review with meta-analysis: Faecal microbiota transplantation for the induction of remission for active ulcerative colitisAlimentary Pharmacology & Therapeutics 2017 46(3):213-24.10.1111/apt.1417328612983 [Google Scholar] [CrossRef] [PubMed]
[84]. Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trialGastroenterology 2015 149(1):102-09.10.1053/j.gastro.2015.04.00125857665 [Google Scholar] [CrossRef] [PubMed]
[85]. Johnsen PH, Hilpüsch F, Cavanagh JP, Leikanger IS, Kolstad C, Valle PC, Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trialThe Lancet Gastroenterology & Hepatology 2018 3(1):17-24.25857665 [Google Scholar] [CrossRef]
[86]. Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trialHepatology 2017 66(6):1727-38.2585766528586116 [Google Scholar] [CrossRef] [PubMed]
[87]. Kootte RS, Levin E, Salojärvi J, Smits LP, Hartstra AV, Udayappan SD, Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota compositionCell Metabolism 2017 26(4):611-19.10.1016/j.cmet.2017.09.00828978426 [Google Scholar] [CrossRef] [PubMed]
[88]. Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label studyMicrobiome 2017 5(1):1010.1186/s40168-016-0225-728122648 [Google Scholar] [CrossRef] [PubMed]
[89]. Zhou YJ, Zhao DD, Liu H, Chen HT, Li JJ, Mu XQ, Cancer killers in the human gut microbiota: Diverse phylogeny and broad spectraOncotarget 2017 8(30):4957410.18632/oncotarget.1731928484095 [Google Scholar] [CrossRef] [PubMed]
[90]. Hu Y, Le Leu RK, Christophersen CT, Somashekar R, Conlon MA, Meng XQ, Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in ratsCarcinogenesis 2016 37(4):366-75.10.1093/carcin/bgw01926905582 [Google Scholar] [CrossRef] [PubMed]