The Gram-negative bacteria of family Enterobacteriaceae, particularly E. coli and Klebsiella species are common pathogens in the hospital setting [1,2]. Over the last two decades these pathogens have emerged as major MDR organisms acquiring resistance via genes borne on and transmitted via plasmids. Carbapenems are considered as one of the agents of last resort for therapeutic management of MDR E. coli and Klebsiella spp, including Extended spectrum beta lactamase (ESBL) producers. The emergence and increasing spread of Carbapenem Resistant Enterobacteriaceae (CRE) is alarming and worrisome. CRE are a serious challenge both clinically especially in the seriously ill as well as with respect to controlling hospital spread.
Carbapenem resistance in Enterobacteriaceae is mainly attributed to the plasmid mediated production of carbapenemase enzymes [3]. Other mechanisms resulting in decreased drug uptake by porin loss or overexpressed efflux pumps or due to overproduction of beta-lactamases AmpC or ESBL in combination with a porin loss, are of less significance in Enterobacteriaceae [4] .
Among Enterobacteriaceae, the KPC, NDM, IMP, VIM and OXA-48-like are the commonly encountered carbapenemases worldwide. There is a distinct geographical distribution of the enzymes in different parts of the world. In India, infections with NDM producing organisms predominate followed by OXA-48-like, while VIM and KPC producing are seen more sporadically [5].
It is imperative for a diagnostic laboratory to accurately and promptly identify a carbapenemase producing organism to facilitate timely and appropriate patient management and trigger hospital infection control protocols.
The detection of carbapenemase encoding genes may be considered as the gold standard for detecting molecular mechanism of carbapenem resistance. Polymerase chain reactions, especially those multiplexed for detection of the major carbapenemases (KPC, NDM, IMP, VIM and OXA-48-like) are commonly used. The commercially available Xpert® Carba-R is a qualitative, real-time PCR test for the detection and identification of these five genes in less than an hour with a sensitivity and specificity of 96.6% and 98.6% respectively and may be performed by laboratory technicians trained in about 30 minutes during installation of the equipment [5,6]. These genotypic tests are expensive and unsuitable in resource poor and low burden areas catering to a diagnostic need. Moreover, as is the case with resistant gene detection, it may be non-expressive or absent in non-carbapenemase producers.
Phenotypic tests thus take predominance in being cost-effective, easy to perform, rapid and detect expressed resistance- which is the key in an effective infection control program. The CLSI guidelines till 2017 recommended the use of MHT and CarbaNP. However, currently they recommend the CarbaNP and modified carbapenemase in activation method (mCIM). The [Table/Fig-1] summarises the various aspects of these phenotypic tests [7,8].
Chromogenic media though available for phenotypic detection of CRE, are not currently recommended by the CLSI. The CMSC, (CHROMagarTM, Paris, France) is one such commercially available chromogenic medium. This is a modification of the original SuperCARBA medium described by Girlich D and is marketed since 2016 [9,10]. The medium detects carbapenemase as well as identifies the isolate as E. coli, Klebsiella sp. or other carbapenemase producing Enterobacteriaceae [11]. The manufacturers report the medium to be highly sensitive and specific, for the OXA-48-like enzymes which are well known to be weakly expressed and also for low inoculum densities of 10 CFU/mL. Studies conducted so far evaluated it as a screening tool for detection of CPE directly from clinical rectal specimens and compare other contemporary chromogenic media.
The present study includes CRE as identified by disk diffusion to determine the utility of CMSC in detecting CRE and compare its performance against the MHT, CarbaNP and conventional molecular detected carbapenemase encoding genes (KPC, NDM, IMP, VIM and OXA-48-like).
Materials and Methods
This was an observational study approved by the Institutional Review Board (IRB Min No: 9830 Dated 07.01.16) and conducted at Christian Medical College and Hospital, a tertiary care 2600 bedded hospital in Vellore, Tamil Nadu, India. It was done over a period of 18 months from January 2016 to June 2017.
A total of 150 consecutive non-repeat CRE (E. coli and Klebsiella sp.) isolated from urine cultures of hospitalised patients (waiver of consent obtained from IRB as bacterial isolates were used without using patient identifiers) in significant or probably significant counts by semi-quantitative culture method and resistant to meropenem (10μg), imipenem (10μg) and ertapenem (10μg) (Sigma-Aldrich, Pvt., Ltd., India) on routine antimicrobial susceptibility testing by disc diffusion were included [8,12].
Genotypic test: The isolates were subjected to a CRE multiplex PCR using standardised protocol described earlier [13]. Briefly, the DNA extraction was performed using boiling lysis method. The multiplex PCR was performed using suitable primers and appropriate cycling conditions for amplification of five gene targets (blaNDM, blaKPC, blaOXA-48-like, blaIMP and blaVIM) in a single PCR reaction [13-15]. The primers used is shown in [Table/Fig-2].
Primers used in the present study [13-15].
| Primer name | Sequence (5’-3’) | PCR product size (bp) |
|---|
| IMP-FIMP-R | GGAATAGAGTGGCTTAAYTCTCCCAAACYACTASGTTATCT | 232232 |
| VIM-FVIM-R | GATGGTGTTTGGTCGCATACGAATGCGCAGCACCAG | 390390 |
| OXA-48 FOXA-48 R | TATATTGCATTAAGCAAGGGCACACAAATACGCGCTAACC | 800800 |
| KPC-FKPC-R | TATATTGCATTAAGCAAGGGCGGGTTGGACTCAAGACG | 10111011 |
| NDM-FNDM-R | GGTTTGGCGATCTGGTTTTCCGGAATGGCTCATCACGATC | 984984 |
Amplification was carried out in the VeritiTM Thermal Cycler (Applied Biosystem, USA) using the following cycling conditions- Initial denaturation: 95°C for 15 min (1 cycle), denaturation: 94°C for 30 sec, annealing: 59°C for 90 sec (30 cycles), extension: 72°C for 90 sec and final extension: 72°C for 10 min (1 cycle). Agarose gel electrophoresis was then performed to visualise the amplicons.
Phenotypic Test
CHROM agar CMSC: Chromogenic medium used for detection and isolation of CPE (Imported and distributed by Neugen diagnostics, R-Biopharm, Hyderabad, India). The medium was prepared as per manufacturer’s instructions and then stored at 4°C, for up to one month [11]. Quality control strains, carbapenemase producing strain Klebsiellapneumoniae BAA 1705 was used as the positive control and carbapenemase non-producing strain Klebsiellapneumoniae BAA 1706 plus carbapenem susceptible E. coli ATCC 25922 were used as negative controls. Inoculated plates were incubated at 37°C for 18-24 hours. Interpretation was done as per manufacturer’s instructions.
CarbaNP test: The CLSI approved test for detection of CPE is based on the principle of in-vitro hydrolysis of the β-lactam ring of a carbapenem agent (imipenem) by a bacterial lysate which is indicated by a change in the pH indicator. The test was performed and interpreted as per 2017 CLSI guideline [8].
MHT: Carbapenemase produced by test isolates streaked on a lawn culture of ATCC 25922 E. coli, renders the carbapenem inactive and allows carbapenem susceptible control strain ATCC 25922 E. coli to grow along the streak line toward the drug disk. Thus, a characteristic cloverleaf-like pattern is obtained. The test was performed and interpreted as per CLSI 2017 guidelines [8].
Results
A total of 150 consecutive carbapenem resistant isolates were tested comprising of E. coli (n=81) and Klebsiella sp. (n=69). Of these, 108 isolates were positive for carriage of one or more carbapenemase encoding genes when subjected to the CRE multiplex PCR. The genes detected are listed in [Table/Fig-3] and the gel electrophoresis picture is shown [Table/Fig-4].
Results of CRE multiplex PCR.
| Gene detected (n=108) | No. of isolates |
|---|
| NDM | 56 |
| OXA-48-like | 34 |
| NDM+OXA-48-like | 12 |
| NDM+OXA-48-like + IMP | 01 |
| NDM+OXA-48-like + VIM | 01 |
| NDM+VIM | 01 |
| OXA-48-like+IMP | 01 |
| VIM | 02 |
Gel electrophoresis image showing DNA ladder, positive controls and test results.
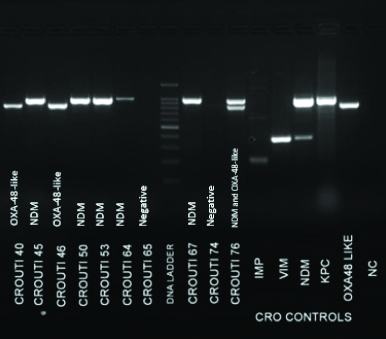
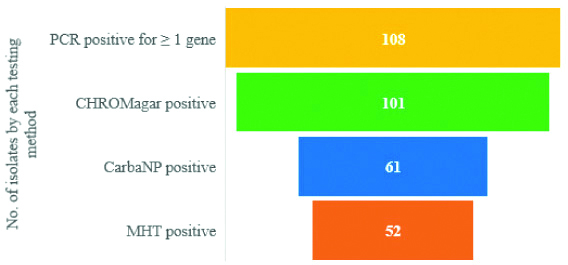
The performance of the three phenotypic tests was then compared to results of the CRE multiplex PCR as shown in [Table/Fig-5]. The results were further analysed based on the ability of each phenotypic test to detect the different classes of carbapenemase enzymes, as summarised in [Table/Fig-6]. Also, the results of the phenotypic tests used are shown in [Table/Fig-7].
Number of carbapenemase producing isolates detected by three phenotypic tests in isolates carrying carbapenemase encoding genes.
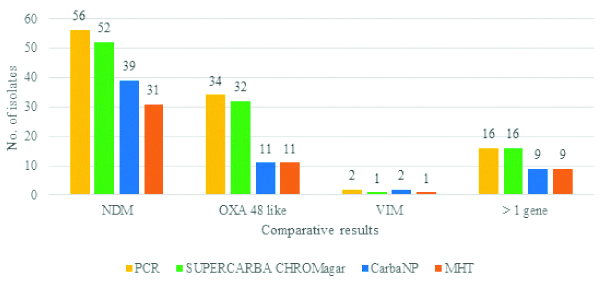
Comparison of performance of phenotypic tests with respect to the carbapenemase encoding gene detected by the multiplex CRE PCR.
Phenotypic test results, a) Modified Hodge test; b) Carba NP and c) mSUPERCARBA CHROMagar.
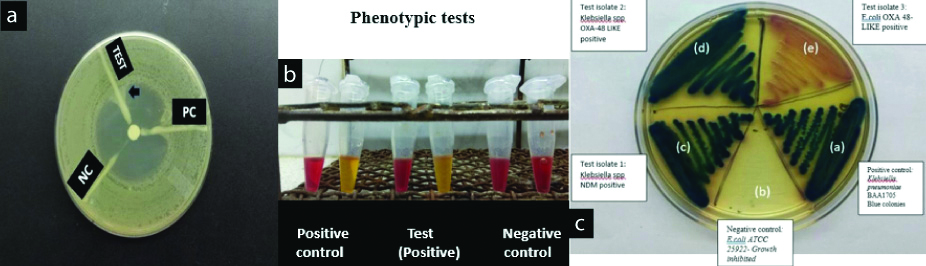
Performance of CMSC media:
After incubation plates were interpreted as follows:
Carbapenemase producing E. coli: dark pink to reddish colonies;
Carbapenemase producing Klebsiella sp.: metallic blue colonies;
Other Gram negative Carbapenemase producing Enterobacteriaceae: colourless colonies or natural pigment seen
Non-carbapenemase producing E. coli/Klebsiella sp.: inhibited
CMSC detected carbapenemase production in 93.5% of isolates with resistant genes (n=101/108 isolates). The sensitivity of the medium in detecting isolates carrying blaNDM and blaOXA-48 like genes respectively was 94.4% (95% CI, 86.2-98.4) and 96% (95% CI, 86-99.5) respectively. The specificity was found to be 35.5% (95% CI, 21.6-51.9).
Performance of the Carba NP test: Carba NP detected carbapenemase production in 56.5% of isolates with resistant gene (n=61/108 isolates). The sensitivity for blaNDM producers and blaOXA-48 like producers respectively was 67.6% (95% CI, 55.5-78.2) and 40.8% (95% CI, 27-55.8), respectively. The specificity of the test was found to be 78.6% (95% CI, 63.2-89.7).
Performance of MHT: MHT detected carbapenemase production in 48% of isolates with resistant genes (n= 52/108 isolates). MHT sensitivity for NDM and OXA-48-like producers respectively was 56.34% (95% CI, 44.0-68.0) and 38.78% (95% CI, 25.2- 53.7), respectively. The specificity was found to be 80.95% (95% CI, 69.9-91-4).
Thus, comparing the performance of the 3 phenotypic tests versus gold standard multiplex CRE-PCR, we determined the diagnostic indicators as highlighted in [Table/Fig-8].
Predicting PCR positivity using phenotypic tests.
| Phenotypic test (n=150) | Sensitivity % | Specificity % | PPV % | NPV % | PLR | NLR |
|---|
| CMSC | 92.5 (87.1-97.3) | 35.5 (21.6- 51.9) | 78.9 (74.8-82.5) | 68.2 (48.5-83) | 1.45 (1.15-1.83) | 0.18 (0.08-0.41) |
| CNP | 56.5 (46.6-66.0) | 78.6 (63.2-89.7) | 87.1 (78.8-92.5) | 41.3 (34.9-47.8) | 2.64 (1.4-4.8) | 0.55 (0.42-0.72) |
| MHT | 48.15 (38.4-57.9) | 80.95 (69.9-91-4) | 86.7 (77.2-92.6) | 37.8 (32.5-43.4) | 2.53 (1.3-4.88) | 0.64 (0.51-0.81 |
PPV: Positive predictive value; NPV: Negative predictive value; LR: Likelihood ratio; CNP: CarbaNP; CMSC: CHROMagarTMmSuperCARBATM
For 42 of the 150 isolates tested, the presence of carbapenemase encoding genes was not detected by the multiplex CRE PCR. Among these, the chromogenic medium, the CarbaNP and the MHT detected the carbapenemase production in 27, 9 and 8 isolates respectively.
Overall sensitivity for detection of carbapenem resistance: The overall sensitivity detecting a carbapenem resistant organism as detected by disk diffusion susceptibility test is 85.3%, 46.6% and 40% for CMSC, CarbaNP and MHT respectively.
Discussion
Carbapenem resistance among E. coli and Klebsiella sp. causing UTI was seen in 2.9% of the isolates as identified by the carbapenem disk diffusion test. Of the 150 CRE isolates tested, 72% had genotypic evidence of carbapenemase production.
The blaNDM gene was found in 52% followed by blaOXA-48-like gene in 32% and they emerged as the predominant carbapenemases among the CRE in this study. Similar findings were observed by Kazi M et al., in Mumbai, India [16]. The SENTRY study conducted during 2006-2007 in different Indian cities found the prevalence of NDM and OXA 48-like producers to be 51% and 39%, respectively [17]. Begum N and Shamsuzzaman MS from Bangladesh in the year 2016 reported similar rates of detection of NDM (50%) followed by OXA-48 like (20%) in their study on carbapenem resistant uropathogens [18].
The sensitivity (%) for detecting carbapenemase production in PCR positive isolates using the CMSC, CarbaNP and MHT was 92.5, 56.5 and 48, respectively. Amar M et al., have reported the sensitivity of CMSC as 83% in detecting CPE (including KPC, NDM, OXA-48, VIM and IMI), and Creighton J et al., reported a sensitivity of 93.9% when the medium was challenged with known carbapenemase producing organisms [10,19]. The sensitivities of CarbaNP and MHT have been variably reported. In a study evaluating the Carba NP by Sun K et al., among Enterobacteriaceae (n=197); including non-carbapenemase producers (n=117), and carbapenemase producers (KPC=18, NDM= 46, IMP=11, OXA=51, and 13 strains harboring two different carbapenemase genes) the sensitivity of CarbaNP was reported as 66.2% [20]. While, Madkour LA et al., reported the sensitivity of CarbaNP as 75% in their study on 203 Gram negative isolates of which 92 were carbapenemase producing with a predominance of NDM and KPC carbapenemases [21]. Osterblad M et al., on 61 carbapenemase-producers of class A, class D, MBLs and 111 non carbapenemase producers, found the sensitivity of CarbaNP to be 88% [22].
The sensitivity of MHT was reported as 61% by Doyle D et al., in their study on 142 CPE from the SMART study network which included KPC, NDM, VIM, OXA-48-like, and IMP-producing isolates [23]. While, Sun K et al., found in the aforementioned mixed isolate population, the MHT sensitivity to be 75% [20]. These sensitivities reflect performance in populations inclusive of KPC producing organisms, which is only sporadically found in our geographic region.
Carbapenemases such as the OXA-48-like enzymes have weaker carbapenemase activity as compared to the others [24]. This makes their phenotypic detection challenging. Among the study isolates carrying blaOXA-48-like gene (n=34), the carbapenemase production was detected best by the CMSC (n=32, 94%) as compared to the CarbaNP and the MHT (n=11, 32%, respectively). The CarbaNP test is known to be less sensitive for detecting OXA-48-like enzymes with CLSI stating sensitivity up to 11% for OXA-48 like producers. Tijet N et al., and Mitra S et al., reported false negative results in 94% (n=31/39) and 33% OXA48-like producing organisms, respectively [25,26].
Similarly, the carbapenemase production among the blaNDM carrying isolates (n=56) was detected better by the CMSC (n=52, 93%) followed by CarbaNP (n=39, 70%) and MHT (n=31, 55%). The low sensitivity of MHT for NDM producers is known. Arif B et al., from Pakistan in their study on 100 isolates; including mainly E. coli and Klebsiella sp., 92 of them being NDM producers, found the sensitivity of MHT to be 42.8% [27].
Conclusively, in a geographic region with a higher burden of blaNDM and blaOXA-48like, phenotypic tests such as MHT or CarbaNP may lead to very major errors in reporting.
The ability of the three tests to rule out carbapenemase gene carriage by the study isolates was analysed using the negative likelihood ratio. The negative likelihood ratio (%) was 0.18 for CMSC, as compared to that of CarbaNP (0.55) and MHT (0.64). Chromogenic CMSC emerged as the superior test to detect isolates not carrying any carbapenemase encoding genes.
Of the 150 isolates tested, 42 were not found to carry the genes tested. Among them, 27 isolates showed growth on the CMSC. This could possibly be due to production of carbapenemases encoded by genes not included in the CRE multiplex PCR. As the medium was challenged by well characterised CRE, it may be proposed that CMSC detects carbapenem resistance conferred by other mechanisms as well. Thus, overall the sensitivity of this media in detecting CRE is 85.3% (n=126/150), making it a good screening test. The CarbaNP and MHT in comparison had overall sensitivities of 46.6 and 40%. Creighton J et al., have also suggested the use of CMSC as a screening test especially in areas with high prevalence of MDR or in situations of outbreaks caused by carbapenemase producing organisms allowing detection in 24 hours of specimen receipt [19].
The faster detection of a CRE is crucial for patient management and in transmission control. While all phenotypic tests may be applied following growth in culture, CMSC may alternatively be also applied as screening media resulting CRE detection in 24 hours. The chromogenic media is simple to prepare and may be used for inoculation of multiple specimens on a single plate. It allows simultaneous identification of E. coli and Klebsiella sp, the two most common CRE pathogens and has a low limit of detection of 101 to 102 CFU/ml [18]. At a cost of 1.2 USD per plate (20ml), it is an ideal direct specimen screening media even in low resource settings.
As highlighted earlier, Carba NP test does have a fast Turnaround Time (TAT) of 2 hours following culture growth and requires minimal technical skill but costs about 3.7 USD. The use of commercially available extraction buffer B-PER II for preparing bacterial extracts may contribute to the slightly higher cost.
To overcome the limitations of the MHT and CarbaNP, the CLSI has now introduced the mCIM test with improved sensitivities especially for the detection of blaNDM and blaOXA-48 like genes [8]. However, its biggest disadvantage is the TAT of 48 hours following culture growth, a total of 3 days after specimen receipt.
Thus, incorporation of CMSC in CRE detection augments earlier detection and implementation of transmission prevention protocols and at much lower costs as compared to other methods.
Limitation
This study was undertaken to determine the utility of the cost-effective CMSC medium to indicate a likely resistant gene carriage as compared to the other CLSI recommended phenotypic tests to confirm the presence of a CRE from culture. Further, the isolates that were CHROMagar positive, but PCR negative were not further characterised to determine the mechanism of resistance.
Conclusion
In the present study on CRE uro-pathogens, E. coli and Klebsiella sp., CMSC emerged as the most sensitive test in detecting blaNDM and blaOXA-48-like gene carriage with an excellent negative likelihood ratio. It correlates well with the disk diffusion screen test, is inexpensive, and has a fast TAT making it an ideal direct screen test in this era of escalating carbapenem resistance.
*EDTA modified CIM
PPV: Positive predictive value; NPV: Negative predictive value; LR: Likelihood ratio; CNP: CarbaNP; CMSC: CHROMagarTMmSuperCARBATM