Materials and Methods
The present descriptive cross-sectional research was conducted for 11 months from January 2017 to November 2017 on 93 E.cloacae isolates obtained from urine, wound, blood, trachea, sputum, CSF, BAL (Broncho alveolar lavage), and catheter in the teaching hospitals of city of Kermanshah, Iran. The study protocol was approved by ethics committee of Kermanshah University of Medical Sciences (No.: 1395.621). In this study, informed consent was obtained from the patients before sample collection. Only E.cloacae present in the clinical specimens of inpatient were evaluated and other Enterobacter species and environmental samples were excluded.
The procured samples were cultured on the special media of Eosin Methylene Blue (EMB) agar and MacConkey agar (Merck, Germany), in sterile conditions [2]. Then; specific tests like culturing in IMVIC and TSI, Simmons’ citrate agar (Himedia CO, India), and indole test were used for identification of E. cloacae. Antibiotic susceptibilities were also tested via Kirby-Bauer method on Mueller-Hinton agar (Himedia CO, India), according to Clinical and Laboratory Standards Institute (CLSI) guideline using 12 discs of antibiotics (MAST, U.K.), namely Cefepime (30 μg), Cefpodoxime (30 μg), Cefotaxime (30 μg), Tetracycline (10 μg), Colistin (10 μg), Ceftazidime (30 μg), Chloramphenicol (10 μg), Aztreonam (30 μg), Co-trimoxazole (25 μg), Tobramycin (10 μg), Imipenem (10 μg), and Amoxicillin/Clavulanic acid (20/10 μg) [12].
The standard strain of Escherichia coli ATCC 25922 was also used as quality control for antibiogram test. E.cloacae isolates, with resistance to three or more antibiotic strains, were further taken into account as MDR strains. Isolates with inhibition zone diameter of approximately 22 mm for Ceftazidime or Cefotaxime were correspondingly investigated for the presence of β-lactamase genes. In addition, ESBL production was confirmed via DDST or Phenotypic Confirmatory Test (PCT). Following the isolation of E.cloacae on the Muller Hinton Agar (Merck, Germany), Ceftazidime, Ceftazidime/Clavulanic acid, Cefotaxime, and Cefotaxime/Clavulanic acid (MAST, UK) discs placed. Following 24 hours incubation at 37°C, the strain with zone of inhibition greater than or equal to 5 mm with clavulanic acid, were designated as an ESBL-producing strain of E.cloacae, as per CLSI guidelines.
The standard strain of Klebsiella pneumoniae ATCC 700603 was also used as the quality control for ESBL-producing strains. Additionally, boiling method for PCR was utilised to extract chromosomal DNA of the isolates. Using specific primers (TakapouZist Co., Iran); the frequency of CTX-M, SHV, and TEM genes was investigated [Table/Fig-1] [13,14]. K.pneumoniae ATCC 7881 containing CTX-M, TEM and SHV genes was used as the positive controls. Correspondingly, PCR was carried out individually for all the genes with total volume of 25 μL constituting of 12.5 μL master mix (Sinaclon Co., Iran), 1 μL of each primer, 3 μL DNA and volume was made up with sterile distilled water. The cycling condition of PCR for all three genes consisted of initial denaturation at 94°C for 5 minutes; 35 cycles of denaturation 94°C for 1 minutes, and annealing temperatures at primer-specific temperature, extension at 72°C for 1 minutes followed by a final extension at 72°C for 7 minutes. The amplified PCR products were separated by agarose gel electrophoresis, and the gel was visualised using gel documentation system (UVP, Germany).
The nucleotide sequence and annealing temperature of primers used for the detection of ESBL-producing E.cloacae.
| Gene | Nucleotide Sequence (5’-3’) | Annealing | Product size (bp) |
|---|
| blaCTX-M-FblaCTX-M-R | 5’-ACGCTGTTGTTAGGAAGTG-3’5’-TTGAGGCTGGGTGAAGT-3’ | 58°C, 45 s | 759 |
| blaTEM-FblaTEM-R | 5’-ATG AGT ATT CAA CAT TTC CG-3’5’-CCA ATG CTT AAT CAG TGA GG-3’ | 55°C, 60 s | 850 |
| blaSHV-FblaSHV-R | 5’-AAG ATC CAC TAT CGC CAG CAG-3’5’-ATT CAG TTC CGT TTC CCA GCG G-3’ | 55°C, 30 s | 231 |
Statistical Analysis
All statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) software version 20 and Chi-square and Fisher’s-tests were used to assess the association between parameters.
Results
The prevalence rate of 93 E.cloacae isolates in males and females was 57 (61.3%) and 36 (38.7%); respectively. The age range was 7 to 81 years with mean age 40.70±19.21 years. The frequency of the isolates in the samples was as follow: urine, 33.4% (31 isolates); wound, 19.3% (18 isolates); blood, 17.2% (16 isolates); trachea, 9.7% (9 isolates); sputum, 7.5% (7 isolates); CSF, 5.4% (5 isolates); BAL, 4.3% (4 isolates); and catheter, 3.2% (3 isolates). Maximum number of isolates were resistant to Amoxicillin/Clavulanic acid (97.8%) and Tetracycline and Ceftazidime (60.2%); whereas Colistin had the highest sensitivity (100%) [Table/Fig-2].
Distribution of E.cloacae strains isolated from clinical specimens.
| Antibiotic | E.cloacae isolated |
|---|
| Resistant | Intermediate | Susceptible |
|---|
| Cefotaxime | 51 (54.8) | 11 (11.8) | 31 (33.4) |
| Cefepime | 38 (40.9) | 11 (11.8) | 44 (47.3) |
| Ceftazidime | 56 (60.2) | 11 (11.8) | 26 (28) |
| Cefpodoxime | 41 (44.1) | 13 (14) | 39 (41.9) |
| Aztreonam | 45 (48.4) | 10 (10.8) | 38 (40.9) |
| Imipenem | 10 (10.8) | 9 (9.7) | 74 (79.5) |
| Amoxicillin-Clavulanate | 91 (97.8) | 1 (1.1) | 1 (1.1) |
| Tobramycin | 35 (37.6) | 8 (8.6) | 50 (53.8) |
| Tetracycline | 56 (60.2) | 4 (4.3) | 33 (35.5) |
| Co-trimoxazole | 48 (51.6) | 9 (9.7) | 36 (38.7) |
| Chloramphenicol | 24 (25.8) | 5 (5.4) | 64 (68.8) |
| Colistin | 0 | 0 | 93 (100) |
Prevalence percentages in parentheses
Out of the 93 isolates, 65 isolates (69.9%) were MDR. According to DDST, 55 isolates (59.1%) were phenotypically ESBL positive. Molecularly, CTX-M was the most frequent gene in ESBL positive isolates with a frequency of 26 (47.3%). The frequency of TEM and SHV genes was also reported by 12 (21.8%) and 3 (5.5%), respectively; and three isolates had CTX-M+TEM genes at the same time. Among MDR isolates, the highest frequency of ESBL genes was observed in urine (13 isolates) and in wound (12 isolates) [Table/Fig-3].
Frequency of ESBL genes in isolates of MDR E.cloacae.
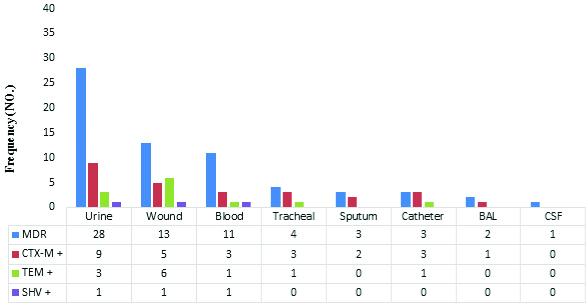
CTX-M was observed to significantly resistant to Cefotaxime (p-value=0.038), Ceftazidime (p-value=0.046), and Imipenem (p-value=0.016), and TEM gene was resistant to Ceftazidime (p-value=0.024) and Chloramphenicol (p-value=0.019) [Table/Fig-4].
Association between resistance to antibiotics and the presence of ESBL genes in isolates of the E.cloacae.
| Genes antibiotic | CTX-M (n=26) | TEM (n=12) | SHV (n=3) |
|---|
| Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible |
|---|
| Cefotaxime | *18 | 2 | 6 | 7 | 0 | 5 | 3 | 0 | 0 |
| Cefepime | 15 | 4 | 6 | 6 | 1 | 5 | 3 | 0 | 0 |
| Ceftazidime | *18 | 3 | 5 | *9 | 1 | 2 | 0 | 0 | 3 |
| Cefpodoxime | 16 | 5 | 5 | 7 | 0 | 5 | 2 | 0 | 1 |
| Aztreonam | 13 | 6 | 7 | 4 | 3 | 5 | 2 | 0 | 1 |
| Imipenem | *19 | 1 | 6 | 7 | 0 | 5 | 1 | 0 | 2 |
| Amoxicillin-Clavulanate | 26 | 0 | 0 | 12 | 0 | 0 | 3 | 0 | 0 |
| Tobramycin | 17 | 2 | 7 | 7 | 2 | 3 | 1 | 0 | 2 |
| Tetracycline | 17 | 2 | 7 | 9 | 2 | 1 | 3 | 0 | 0 |
| Co-trimoxazole | 13 | 5 | 8 | 8 | 1 | 3 | 2 | 0 | 1 |
| Chloramphenicol | 17 | 4 | 5 | *9 | 0 | 3 | 2 | 0 | 1 |
| Colistin | 0 | 0 | 26 | 0 | 0 | 12 | 0 | 0 | 3 |
| Significant p-value=* |
Chi-square and Fisher’s tests; *p<0.05 statistically significant
CTX-M: Cefotaximase-Munich, SHV: Sulphydrylvariable, TEM: Temoneira
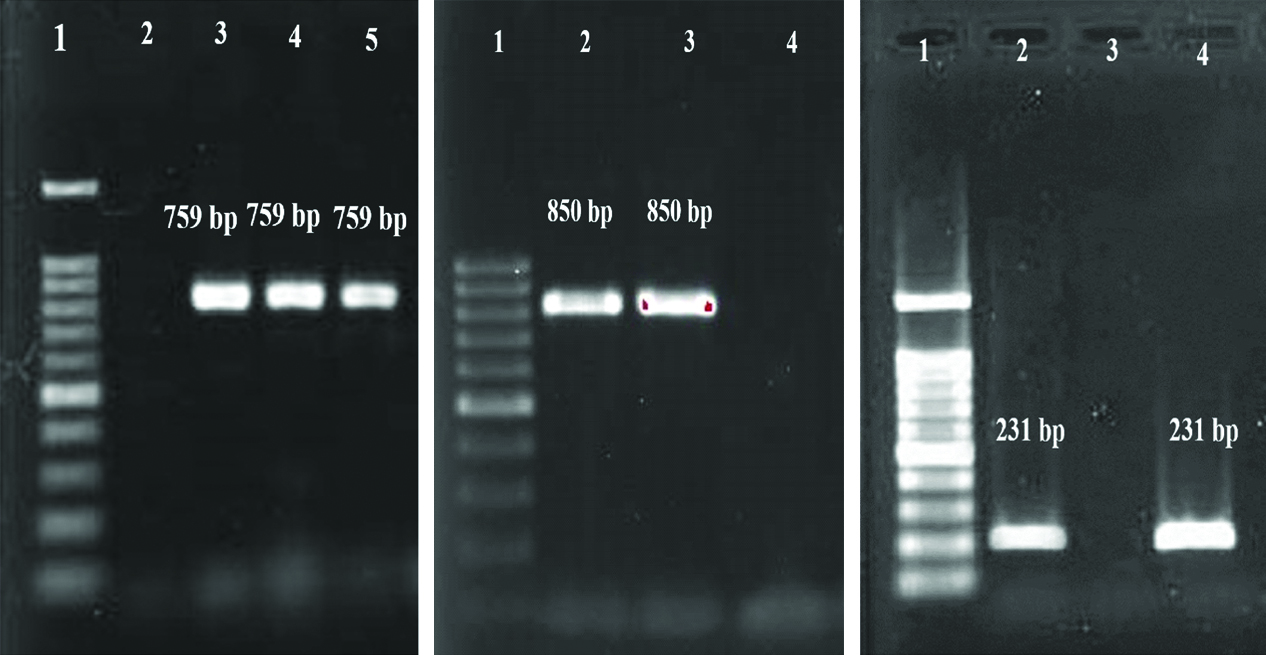
Upon PCR, 759 bp bands were obtained for CTX-M gene, 850 bp for TEM gene and 231 bp for SHV gene [Table/Fig-5].
Gel electrophoresis of PCR products of ESBL gene.
(a) CTX-M: 1- Ladder (100 bp), 2- Negative control, 3- Positive control (759 bp), 4,5- Positive sample (759 bp),
(b) TEM: 1- Ladder (100 bp), 2-Positive control (850 bp), 3-Positive sample (850 bp) 4- Negative control, and
(c) SHV: 1- Ladder (100 bp), 2- Positive control (231 bp), 3- Negative control, 4- Positive sample (231 bp).
Discussion
Enterobacter species are hospital acquired causative agents of nosocomial infections. ESBL-producing isolates are a major concern because of their resistance to various classes of antibiotics as well as their infectious nature [15]. In contrast to published studies, in the present study the maximum number of E.cloacae isolates was found in urine [2,16]. Maximum number of E.cloacae isolates was observed to be resistant to Amoxicillin/Clavulanate; and these findings were similar to the results of investigations in the cities of Tehran and Qazvin, Iran [17]. Cephalosporins are among the most commonly used antibiotics for treating bacterial infections, and in the present study, 50% E.cloacae isolates were resistant to different Cephalosporins with highest resistance to Ceftazidime (60%). Similar results were reported by Malekzadegan Y et al., where the maximum number of isolates was resistant to Cefotaxime [16]. In another Germany based study, 40% of the E.cloacae strains were reported to be resistant to broad-spectrum Cephalosporins [10].
In the present study, most susceptibilities were reported to Colistin and Imipenem. In the investigation by Ghorashi Z et al., Colistin and Imipenem were also introduced as the most effective antibiotics [18]. Thus, suggesting that some antibiotics are still useable for effective treatment of these bacterial infections. However, a Jordan based study reported that E.cloacae isolates were resistant to Carbapenems, including Meropenem and Imipenem [19]. These discrepanies in the results of drug resistance in multiple studies might be because of difference in bacterial colony as well as their prevalence in hospital environments, drug resistance gene distribution, variance in antibiotic usage pattern, and their administration in patients.
According to the susceptibility antibiotic test, 69.9% (65 isolates) of the isolates were found to be MDR. In another study from the same region; 54 (75%) of the isolates were reported to be MDR isolates, thus confirming the results of the present study [2]. In another Iran-based study, prevalence of MDR was between 26.7% and 91.8%, and the wide range of resistance was attributed to misuse of broad-spectrum antibiotics in hospitals [16,17]. Considering E.cloacae, the main mechanism of drug resistance is production of ESBL. From 93 isolates of E.cloacae, 55 (59.1%) were phenotypically positive for ESBL. In contrast to our results, few Iran based studies reported 44.2-100% prevalence rate of ESBL producing Enterobacter isolates [16,17,20]. In other countries, such as Brazil, Algeria and India, the prevalence rate of ESBL-producing Enterobacter was also reported to be 20%, 47.6% and 73.4% respectively [21-23]. The difference in the results of the ESBL-producing Enterobacter isolates can be attributed to diversity in the consumption pattern of various antibiotics, especially ESBL, and implementation of appropriate strategies for management of hospital based infections in treatment centres.
Prior research suggests that CTX-M, TEM, and SHV are the most common ESBL genotypes in E.cloacae strains. In the present study, CTX-M gene was found to have highest frequency (47.3%) followed by TEM (21.8%) and SHV (5.5%) genes.
In the study by Ramazanzadeh R et al., Enterobacter isolates obtained from ICUs contained only the CTX-M gene [13]. In another study by Ghanavati R et al., TEM and CTX-M genes were presented as the most commonly reported genes [20]. In different studies from other countries such as Germany, Algeria, and Jordan; CTX-M was reported to be the most frequent gene [10,19,24]. In the study by Bogner C et al., on ESBL positive and third-generation cephalosporins resistant strains of K. pneumoniae, E. coli, K. oxytoca, and E.cloacae, CTX-M gene had highest frequency followed by SHV and TEM genes [25]. These findings were contradictory to Moosavian M et al., study on clinical isolates of Enterobacteriaceae, where SHV and TEM were the most frequent genes and no CTX-M gene was reported [26]. These variations in the prevalence of ESBLs can be due to differences in the type and the volume of antibiotics used and differences in the collection time of isolates.
In the current study, 3 isolates were observed to have 2 genes (CTX-M+TEM). The simultaneous presence of ESBL genes in a single isolate had been also reported in other studies [19,27]. The presence of combination of genes confines treatment choices and raise challenges in true diagnosis [28]. Since these genes are coded by the plasmids, they can easily transfer in hospital environments; via risk factors such as antibiotic usage, catheter, and person-to-person contact. In the present study, significant association was observed between presence of CTX-M gene and resistance to third-generation Cephalosporin, and the presence of TEM gene and resistance to Ceftazidime. Liu J et al., study showed a significant relationship between antibiotic resistance in E.cloacae and presence of ESBL genes [29]. These findings suggest the importance of molecular diagnosis for determining the treatment plan.
Limitation
The lack of access to the patient information, and other potential resistant β-lactamase in isolates of E.cloacae were not identified. E.cloacae isolates should be further examined to determine the frequency of genes resistant to quinolones, aminoglycosides and their relationship to drug resistance patterns.
Conclusion
The presence of CTX-M, TEM, and SHV genes in E.cloacae indicates their resistance to β-lactam antibiotics. Broad-spectrum β-lactam antibiotics are widely used for treating the gram-negative bacterial infection; however, over the past decade the resistance to this class of antibiotics have increased significantly. This rise in antibiotic-resistant in E. cloacae isolates is a serious concern for public health. Therefore, administration of antibiotics should be closely monitored, and both phenotypic and molecular diagnostic techniques should be used to identify the type of genetic resistance to develop effective treatments to prevent emergence of antimicrobial-resistance.
Funding: The present study was supported by Kermanshah University of Medical Sciences, Kermanshah, Iran.
Prevalence percentages in parentheses
Chi-square and Fisher’s tests; *p<0.05 statistically significant
CTX-M: Cefotaximase-Munich, SHV: Sulphydrylvariable, TEM: Temoneira