Prevalence of blaCTX-M, blaCTX-M-2, blaCTX-M-8, blaCTX-M-25 and blaCTX-M-3 Genes in Escherichia coli Isolated from Urinary Tract Infection in Kermanshah City, Iran
Alisha Akya1, Mahnaz Ahmadi2, Sepideh Khodamoradi3, Mohammad Reza Rezaei4, Nahid Karani5, Azam Elahi6, Roya Chegene Lorestani7, Mansour Rezaei8
1 Associate Professor, Department of Medical Microbiology, Infectious Diseases Research Center, Kermanshah University of Medical Sciences, Kermanshah, Iran.
2 MSc in Medical Surgical Nursing, Imam Reza Hospital, Kermanshah University of Medical Sciences, Kermanshah, Iran.
3 PhD, Department of Biology, Faculty of Basic Sciences, Islamic Azad University, Shahr-e-Qods Branch, Tehran, Iran.
4 Associate Professor, Department of Emergency Medicine, School of Medicine, Kermanshah University of Medical Sciences, Kermanshah, Iran.
5 BSc in Nursing, Imam Reza Hospital, Kermanshah University of Medical Sciences, Kermanshah, Iran.
6 MSc in Medical Microbiology, Infectious Diseases Research Center, Kermanshah University of Medical Sciences, Kermanshah, Iran.
7 MSc in Medical Microbiology, Infectious Diseases Research Center, Kermanshah University of Medical Sciences, Kermanshah, Iran.
8 Associate Professor, Department of Biostatistics, School of Health, Kermanshah University of Medical Sciences, Kermanshah, Iran.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Mansour Rezaei, School of Health, Kermanshah University of Medical Sciences, Kermanshah, Iran.
E-mail: mansourreza05@gmail.com
Introduction
Urinary Tract Infection (UTI) is one of the most common bacterial infections and Escherichia coli is the most common organism that causes UTI. However, the incidence of community acquired UTI caused by Extended spectrum beta-lactamase (ESBL)-producing strains of E.coli, in particular CTX-M genes, is on the rise worldwide.
Aim
To detect the frequency of CTX-M gene subgroups in uropathogenic E.coli.
Materials and Methods
In this descriptive-analytical study, 240 isolates of E. coli were studied. All isolates were isolated from UTIs in Kermanshah University of Medical Sciences, Kermanshah, Iran, in 2014 to 2015. After screening for ESBL, the CTX-M, CTX-M-2, CTX-M-8, CTX-M-25 and CTX-M-3 genes were detected among ESBL- producing isolates using PCR.
Results
Of the 240 E. coli isolates, 67 were ESBL-producing isolates. Sixty one isolates (91%) contained CTX-M gene, of which 57 (85%), 3 (4.5%), 3 (4.5%) and 1(1.49%) contained CTX-M-3, CTX-M-8, CTX- M-25 and CTX-M-2, respectively.
Conclusion
Due to the high resistance of E. coli to beta-lactam drugs in this region, these drugs have limited effects for treatment of UTI in outpatient. The frequency of CTX-M-2, CTX-M-8,CTX-M-25 beta-lactamases in isolates of E. coli is relatively low but the overall prevalence of CTX-M and CTX-M-3 beta-lactamases is high which indicates the spread of drug resistance.
Introduction
UTI is one of the most common bacterial infections [1]. Failure to diagnose and improper treatment of UTI may lead to complications such as kidney damage and hypertension [2]. Among the uropathogenic bacteria in the outpatients and inpatients, Escherichia coli (E. coli) is the most common organism [1,3]. Given the obvious role of E. coli as the main and prevalent cause of UTIs in all ages, it is important to recognise its regional susceptibility pattern to antibiotics [1]. The sensitivity of bacteria to diverse antibiotics varies in different regions, which can be the consequence for the usage of various types and quantity of antibiotics in each region [3]. The first line of antibiotics for the treatment of UTIs is usually determined experientially; therefore, it is essential to obtain accurate and up-to-date information on the antibiotic susceptibility pattern of the regionally circulating strains. Beta-lactam antibiotics are often used for the treatment of bacterial infections [4]. The main resistance mechanism used in gram-negative bacteria against beta-lactam antibiotics is the production of beta-lactamase enzymes to hydrolyze the beta-lactam ring of antibiotics [5]. E. coli strains produce ESBLs resistant to beta-lactam antibiotics [4].
The common types of ESBLs in gram negative bacteria are TEM, SHV and CTX-M. In recent years, the CTX-M group has been increasingly reported in gram-negative bacteria, especially E. coli [6]. Until recently, more than 123 types of CTX-M had been identified and reported [7]. These beta-lactamases have no genetic linkage with TEM and SHV beta-lactamases [8]. In CTX-M, the presence of a serine amino acid at position 237 has led to the expansion of its beta-lactamase activity spectrum [9]. The CTX-M subgroups are divided into 5 main groups based on the amino acid sequences; Group 1 (CTX-M-1, 3, 10, 11, 12, 15, 22, 23, 28, 29, 30, UOE- 1), Group 2 (CTX-M-2, 4, 5, 6, 7, 20, Toho-1), Group 3 (CTX-M-8), Group 4 (CTX-M-9, 13, 14, 16, 17, 19, 21, 27, Toho-2) and Group 5 (CTX-M-25,26) [8].
Over the past decades, CTX-M subgroups have been more frequently reported than TEM and SHV in Europe, North America and Asia [10,11]. Similarly, the reports for CTX-M type have also been in rise in Iran [12]. Given the diversity of CTX-M beta-lactamases and their different effects on susceptibility to various antibiotics, it is epidemiologically essential to determine the frequency of subgroups of these resistant genes in E. coli. Previous studies in Iran primarly have focused on the main ESBL groups in clinical strains of E. coli, while various CTX-M subtypes have been less investigated in different regions of Iran [2,3,12]. The first part of data derived from this work has already been published which indicate the high frequency of blaCTX-M1, blaCTX-M14 and blaCTX-M15 among isolates in our region [13]. Therefore, the present study aimed to evaluate the frequency of other blaCTX-M subtyped genes including CTX-M-2, CTX-M-8, CTX-M-25 and CTX-M-3 in E. coli isolates.
Materials and Methods
Isolation of Bacteria and Collection of Sample Data
The present descriptive-analytic study was conducted on 240 isolates of E. coli obtained from outpatients with UTI referred to the Clinic of Kermanshah University of Medical Sciences and the Central laboratory. The study was approved by the Kermanshah University Ethics Committee (Approval number: IR.KUMS.REC.1393.519). All patients agreed to participate in the study and signed the informed consent form. The UTI was defined as the presence of 105 or more E. coli bacteria per ml of midstream urine sample in the patients who were clinically suspected to UTI [14]. The patients’ data including age, gender and type of samples were also collected. E. coli isolates from urine samples were isolated and identified using Gram staining, morphology, culture characteristics and conventional biochemical tests for verification, including oxidase, simmons’ citrate, urease, phenylalanine deaminase, lysine decarboxylase, Sulfur Indole Motility Medium (SIM), Triple Suger Iron Agar (TSI), Methyl Red/Voges-Proskauer (MR/VP) [15].
Screening Isolates for ESBL and their Resistance Pattern
The verification tests for analysing the phenotypes were used to screen ESBL production in bacteria. This method requires combination discs including ceftazidime (30 μg) + clavulanic acid (10 μg) and cefotaxime (30 μg) + clavulanic acid (10 μg) (MAST, England). The diameter of the bacterial inhibition zone around the combination disc about 5 mm or more than the inhibition zone diameter of the single disc relating to the same antibiotic was considered as the ESBL-producing isolates. The standard strain of E. coli ATCC 35218 was used for qualitative control of ESBL-producing isolates. The ESBL-positive isolates were further tested using PCR for evaluating the frequency of CTX-M, CTX-M-2, CTX-M-8, CTX-M-25, and CTX-M-3 genes. The antibiotic susceptibility of isolates to beta-lactam antibiotics was assessed using Disk diffusion test according to Clinical and Laboratory Standards Institute (CLSI 2015) guidelines and antibacterial discs, including ceftriaxone (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), aztreonam (30 μg), ampicillin (10 μg) and imipenem (10 μg) (MAST, England). The results were interpreted based on standard tables of CLSI [16]. Meanwhile, the standard strain of E. coli ATCC 25922 and ATCC 700603 was used as qualitative control.
Detection of CTX-M Genes
The specific primers using PCR were applied to detect the CTX-M genes and related subgroups of CTX-M, CTX-M-2, CTX-M-8, CTX-M-25 and CTX-M-3 [17-19]. The sequences of used primers and the size of the PCR products have been listed in [Table/Fig-1]. Initially, the isolates were cultured on nutrient agar medium and DNA was extracted using boiling method. The PCR was performed using the reaction solution with final volume of 25 μL, including 12.5μl of Master Mix 2X, 3 μL of DNA template, 10 picomoles (1μL) of each of paired primers, and 7.5 μL of sterilized double-distilled water. The agarose gels were immersed in 0.5 to 1 mg/L of ethidium bromide for 10 minutes, rinsed with distilled water and then examined by Gel-Documentation (BioRad, USA). The DNA sequencing was performed using an ABI 3730XL DNA analyser apparatus (Macrogen Inc., Korea). The DNA sequence data were analysed for homology with genetic data using the National Center for Biotechnology Information GenBank database (http:/www.ncbi.nlm.nih.gov/BLAST/).
| Gene | Forward and reverse primers (5’-3’) | Amplicon size (bp) |
|---|
| blaCTX-M-2 | F:ATGATGACTCAGAGCATTCGR:TTATTGCATCAGAAACCGTG | 884 |
| blaCTX-M-8 | F:ATGATGAGACATCGCGTTAAGR:CGGTGACGATTTTCGCGGCAG | 924 |
| blaCTX-M-25 | F:CACACGAATTGAATGTTCAGR: TCACTCCACATGGTGAG | 864 |
| blaCTX-M-3 | F:AATCACTGCGCCAGTTCACGCTGAACGTR: TTCGTCTCCCAGCTGT | 540-600 |
| blaCTX-M | F:TTTGCGATGTGCAGTACCAGTAAR:CGATATCGTTGGTGGTGCCAT A | 544 |
Statistical Analysis
The collected data were analysed by SPSS 19 using descriptive and analytical statistical indexes.
Results
Data of Samples and Patients
Out of 67(27.9%) ESBL-producing E. coli isolates, 34 (50.7%) belonged to the Clinic of University and 33 (49.3%) to the Central Laboratory. Of these 67 samples, 57 belonged to females (85.1%) and 10 to males (14.1%). The mean age of patients was 43.1±22.1 years with the maximum age of 97 years and the minimum age of 1 year.
The amplification results of target genes are displayed in [Table/Fig-2].
Electrophoresis of PCR products for CTX-M genes.
Row 1: Marker (DNA ladder, 100bp); Row 2: CTX-M-3 (540 bp); Row 3: CTX-M (544 bp); Row 4: CTX-M-8 (924 bp); Row 5: CTX-M-25 (846 bp); Row 6: CTX-M-2 (884 bp); Row 7: Negative control
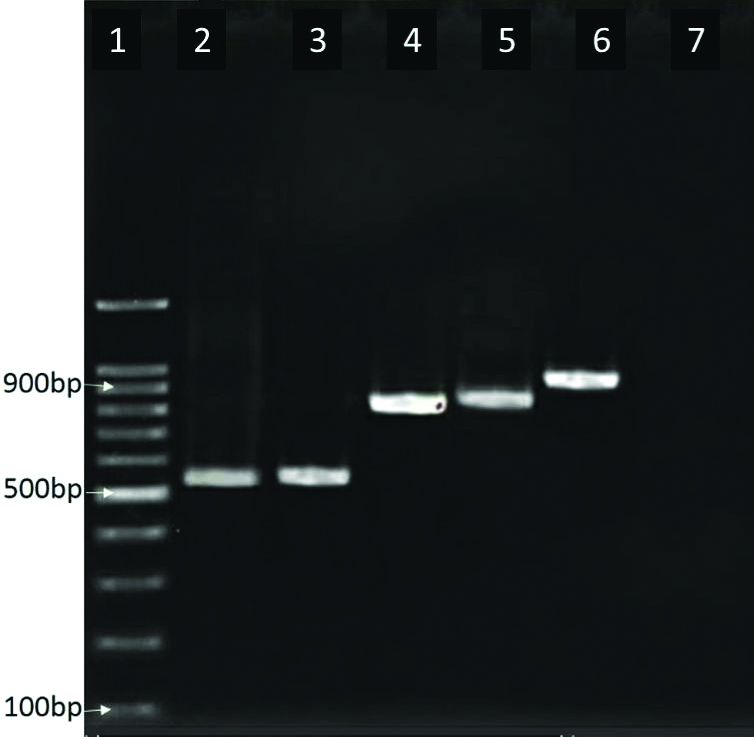
Of the 67 ESBL-producing E. coli isolates, 61 isolates (91%) contained the CTX-M gene, of which 57 (85%) had CTX-M-3 and three (4.5%) had CTX-M-8, three (4.5%) had CTX-M-25 and one (1.49%) contained CTX-M-2.
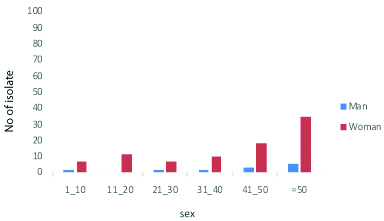
The susceptibility to beta-lactam antibiotics in CTX-M-producing and non CTX-M isolates are listed in [Table/Fig-3]. [Table/Fig-4] shows the frequency distribution of CTX-M-containing isolates based on age and gender.
Comparison the susceptibility to beta-lactam antibiotics among the CTX-M producing and non CTX-M isolates of E. coli.
| Antibiotics | CTX-M producer | Non-CTX-M | Chi-square | p-value |
|---|
| Ampicillin | 1 (1.6%) | 34 (19%) | 11 | 0.001 |
| Ceftriaxone | 1 (1.6%) | 164 (91.6%) | 171.5 | 0.001< |
| Cefotaxime | 2 (3.3%) | 164 (91.6%) | 166.5 | 0.001< |
| Ceftazidime | 13 (21.3%) | 167 (93.3%) | 125.7 | 0.001< |
| Aztreonam | 6 (9.8%) | 166 (92.7%) | 154 | 0.001< |
| Imipenem | 61 (100%) | 179 (100%) | - | - |
Distribution of CTX-M isolates by age and sex.
Discussion
The outpatients reffering from UTI are primarily treated empirically which requires the awareness of antibiotic susceptibility and the frequency of antibiotic resistance genes in each country or region [2]. Beta-lactams are usually used against gram-negative bacteria such as E. coli [5]. The results of the present study indicated a high resistance to beta lactam antibiotics among E. coli isolates. Accordingly, ampicillin resistance was observed to be very high in these isolates, which is consistent with the results of other studies conducted in Iran and other countries [2,20,21]. These findings indicate the spread of resistance to this drug. On the other hand, the present results revealed all E. coli isolates were sensitive to imipenem that is consistent with the results of other studies [2,20-23].
Comparison of the resistance pattern of isolates showed that the antibiotic resistance of CTX-M-producing isolates was much higher than non CTX-M isolates, emphasizing the important role of CTX-Ms for spread of antibiotic resistance in E. coli strains, which needs to be considered in the treatment of UTIs. Compared to the previous studies from other countries, our results indicate an increase in the frequency rate of ESBL-producing E. coli isolates [2,5,10].
Given the results of other studies and the fact that most previous researches in Iran were basically focused on clinical isolates of inpatient which as expected to be more resistant to antibiotics. The high frequency of CTX-M isolates among outpatients in our region suggest the dissemination of this gene group. The beta-lactamase genes in this bacterium, especially the CTX-M genes, are major factors involved in the increasing resistance to beta-lactam antibiotics. Organisms containing these genes can also exacerbate the pathogenicity and increase the mortality in the patients [24].
Studies conducted in different countries including Iran, reported a various frequency rates for the CTX-M genes [24-26]. For example, a study from India (2015-2016) on E. coli isolated from UTI reported a prevalence rate of 82.6% for CTX-M gene in ESBL-producing strains [27]. In another study in Iran (2015), the prevalence of CTX-M was 68.9% in 29 strains of ESBL-producing E. coli [28]. Our findings along with the aforementioned studies indicate that CTX-M-producing E. coli have become more frequent and the production of this beta-lactamase is increasing on the rise. The high frequency of CTX-M genes could be the consequence of over prescription of cephalosporins in our region followed by the transmission of plasmids among strains of bacteria. On the other hand, this gene has the ability of transmission between animal (poultry) and humans bacteria, which is considered as the another source of resistant genes dissemination [29].
It has been shown that CTX-M genes are located upstream to the ISEcp1 conjugated sequences which may explain the expression and transmission of this gene group [30]. In our study, the relatively low percentage of ESBL-producing strains had CTX-M-2, CTX-M-8 and CTX-M-25 genes, which is similar to other research results. For instance, in a study in Iran (2009), only 0.7% of the strains of E. coli had CTX-M-25 and none of the isolates had CTX-M-2 [31]. In another Iranian study in 2013, none of the E. coli isolates contained CTX-M-2 [32]. The results of studies in other countries are also in agreement with the findings of research conducted in Iran. For example, the studies in China (2008) and Spain (2011) reported no production of CTX-M-2, 8, 25 in E. coli [33,34]. Furthermore, in a study in France (2006), none of the isolates had the CTX-M-8 gene, and only one isolate (2.27%) was detected to have CTX-M-2 gene among E. coli isolates [6]. In Egypt (2014), the frequency of CTX-M-2 gene was found to be zero in E. coli isolates and the frequency of CTX-M-25 gene was reported to be 1.3% [4]. Considering the results of the above studies, it seems that the prevalence of these genes in E. coli isolates is still low and varies in different geographic regions.
The prevalence of CTX-M-3 subtype varies in different regions. Studies in Iran on Group 1 of CTX-M have reported different frequencies which ranged from 35.87% to 87.5% [10]. For instance, a study in Rasht (Iran) showed that only 2.1% of the isolates possessed the CTX-M-3. This gene was also observed in 88% of ESBL-positive isolates in France and 84% in India [35,36]. The CTX-M-I was found in 60.6% of ESBL-positive isolates in Poland [37], but CTX-M-3 was reported to have an extremely low prevalence in other countries. For example, studies conducted in Canada reported that the prevalence of CTX-M-3 was 1% in 2007 and 2% in 2009. The prevalence of this gene in Germany (2009) was reported to be 4.7% [8,38]. In the present study, 85% of ESBL-positive isolates exhibited the CTX-M-3 gene, which can demonstrate the spread of this gene among E. coli strains in our region.
Limitation
As a limitation of our study, it was not possible to collect all relevant data of patients’ UTIs including underlined diseases, the history of UTIs and previous treatments.
Conclusion
Due to the high resistance of E. coli to beta-lactam drugs in our region, especially cephalosporins and penicillins, these antibiotics cannot effectively treat UTIs in outpatients. Although the prevalence of CTX-M-2, CTX-M-8 and CTX-M-25 subtypes of beta-lactamases in E. coli in Kermanshah region are relatively low, the prevalence of CTX-M beta-lactamases and CTX-M-3 subtype is high, which indicate the spread of drug resistance among the strains of this bacterium.
[1]. Gupta K, Hooton TM, Stamm WE, Increasing antimicrobial resistance and the management of uncomplicated community-Acquired urinary tract infectionsAnn Intern Med 2001 135:41-50.10.7326/0003-4819-135-1-200107030-0001211434731 [Google Scholar] [CrossRef] [PubMed]
[2]. Mohajeri P, Izadi B, Rezai M, Falahi B, Khademi H, Ebrahimi R, Assessment of the frequency of extended spectrum beta lactamases producing E. coli isolated from urinary tract infections and its antibiotic resistance pattern in KermanshahJ Ardabil Univ Med Sci 2011 11:86-94. [Google Scholar]
[3]. Akya A, Gheisari H, Mohammadi G, Khodadoost M, Study of the pattern of plasmid and antibiotic resistance of E. coli isolated from outpatients with urinary tract infectionSci J Kurdistan Uni Med Science 2015 20:89-96. [Google Scholar]
[4]. El-Naghy WS, Wafy AA, Elfar NN, Taha A, Shahba A, Noor-eldeen NM, Multiplex PCR for detection of bla CTX-M genes among the Extended Spectrum Beta Lactamase (ESBL) producing gram-negative IsolatesEgypt J Med Microbiol 2014 23:107-14.10.12816/0024357 [Google Scholar] [CrossRef]
[5]. Paterson DL, Resistance in gram-negative bacteria: EnterobacteriaceaeAm J Infect Control 2006 34:S20-28.discussion S64-7310.1016/j.ajic.2006.05.23816813978 [Google Scholar] [CrossRef] [PubMed]
[6]. Naas T, Oxacelay C, Nordmann P, Identification of CTX-M-type extended-spectrum-beta-lactamase genes using real-time PCR and pyrosequencingAntimicrob Agents Chemother 2007 51:223-30.10.1128/AAC.00611-0617088478 [Google Scholar] [CrossRef] [PubMed]
[7]. Al Naiemi N, Duim B, Bart A, A CTX-M extended-spectrum beta-lactamase in Pseudomonas aeruginosa and StenotrophomonasmaltophiliaJ Med Microbiol 2006 55:1607-08.10.1099/jmm.0.46704-017030926 [Google Scholar] [CrossRef] [PubMed]
[8]. Pitout JD, Hossain A, Hanson ND, Phenotypic and molecular detection of CTX-M-beta-lactamases produced by Escherichia coli and Klebsiella sppJ Clin Microbiol 2004 42:5715-21.10.1128/JCM.42.12.5715-5721.200415583304 [Google Scholar] [CrossRef] [PubMed]
[9]. Rossolini GM, D’Andrea MM, Mugnaioli C, The spread of CTX-M-type extended-spectrum beta-lactamasesClin Microbiol Infect 2008 14(1):33-41.10.1111/j.1469-0691.2007.01867.x18154526 [Google Scholar] [CrossRef] [PubMed]
[10]. Sidjabat HE, Paterson DL, Adams-Haduch JM, Ewan L, Pasculle AW, Muto CA, Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western PennsylvaniaAntimicrob Agents Chemother 2009 53:4733-39.10.1128/AAC.00533-0919687234 [Google Scholar] [CrossRef] [PubMed]
[11]. Canton R, Coque TM, The CTX-M blactamase pandemicCurr Opin Microbiol 2006 9:466-75.10.1016/j.mib.2006.08.01116942899 [Google Scholar] [CrossRef] [PubMed]
[12]. Sharifi yazdi M, Azarsa M, Shirazi M, Rastegar Lari A, Owlia P, Fallah Mehrabadi J, The frequency of extended spectrum beta lactamase and CTX M-I of escherichia coli isolated from the urine tract infection of patients by phenotypic and PCR methods in the city of Khoy in IranJAMBR 2011 19(77):53-61. [Google Scholar]
[13]. Akya A, Khodadoost M, Prevalence of blaCTX-M1, blaCTX-M14 and blaCTX-M15 genes among Escherichia coli isolated from urinary tract infections in outpatients, Kermanshah city, IranJ Kerman Uni Med Sci 2016 23(2):145-55. [Google Scholar]
[14]. Fauci AS, Harrison’s principles of internal medicine / editors, Anthony S. Fauci ... [et al.] 2008 17th edNew YorkMcGraw-Hill Medical [Google Scholar]
[15]. Winn WC, Koneman EW, Koneman’s color atlas and textbook of diagnostic microbiology 2006 6th edPhiladelphiaLippincott Williams & Wilkins [Google Scholar]
[16]. Clinical and Laboratory Standards Institute (CLSI) guidelines 2015: Performance standards for antimicrobial susceptibility testing; 24th Information supplement; M100-S24; Vol.34 No.1.CLSI, Wayne, PA [Google Scholar]
[17]. Chmelnitsky I, Carmeli Y, Leavitt A, Schwaber MJ, Navon-Venezia S, CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, IsraelAntimicrob Agents Chemother 2005 49:4745-50.10.1128/AAC.49.11.4745-4750.200516251320 [Google Scholar] [CrossRef] [PubMed]
[18]. Ben-Ami R, Rodríguez-Baño J, Arslan H, Pitout JD, Quentin C, Calbo ES, A multinational survey of risk factor for infection with extended-spectrum beta-lactamase-producing entrobacteriaceae in nonhospitalized patientsClin Infect Dis 2009 49:682-90.10.1086/60471319622043 [Google Scholar] [CrossRef] [PubMed]
[19]. Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L, Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitalsAntimicrob Agents Chemother 2003 47:3724-32.10.1128/AAC.47.12.3724-3732.200314638473 [Google Scholar] [CrossRef] [PubMed]
[20]. Mantadakis E, Tsalkidis A, Panopoulou M, Pagkalis S, Tripsianis G, Falagas ME, Antimicrobial susceptibility of pediatric uropathogens in Thrace, GreeceInt Urol Nephrol 2011 43:549-55.10.1007/s11255-010-9768-x20524067 [Google Scholar] [CrossRef] [PubMed]
[21]. Akram M, Shahid M, Khan AU, Etiology and antibiotic resistance patterns of community-acquired urinary tract infections in JNMC Hospital Aligarh, IndiaAnn Clin Microbiol Antimicrob 2007 6:410.1186/1476-0711-6-417378940 [Google Scholar] [CrossRef] [PubMed]
[22]. Shahcheraghi F, Nasiri S, Naviri H, Evalouation of presence the bla-SHV and bla- TEM ß-Lactamase genes in clinical isolates resistant E. coli to antibiotics from Tehran hospitalIran J Med Microbiol 2007 :1-8. [Google Scholar]
[23]. Andrade SS, Sader S, Jones R, Pereira AS, Pignatari AC, Gales AC, Increaced resistance to first-line agents among bacterial pathogens isolated from urinary tract infections in Latin America. time for local guidelines?Men Inst Oswaldo Cruze 2006 101:741-48.10.1590/S0074-0276200600070000617160281 [Google Scholar] [CrossRef] [PubMed]
[24]. Mirzaee M, Pourmand MR, Chitsaz M, Mansouri S, Antibiotic resistance to third generation cephalosporins due to CTX-M-type extended-spectrum beta-lactamases in clinical isolates of escherichia coliIran J Public Health 2009 38:10-17. [Google Scholar]
[25]. Burcu B, Acik L, Sultan N, Phenotypic and molecular characterization of SHV, TEM, CTX-M and extendedspectrum ß-lactamase produced by E. coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospitalAfr J Microbiol Res 2010 4:650-54. [Google Scholar]
[26]. Goyal A, Prasad KN, Prasad A, Gupta S, Ghoshal U, Ayyagari A, Extended spectrum beta-lactamases in E. coli & Klebsiella pneumoniae & associated risk factorsIndian J Med Res 2009 129:695-700. [Google Scholar]
[27]. Jena J, Sahoo RK, Debata NK, Subudhi E, Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum β-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adultes3 Biotech 2017 7(4):24410.1007/s13205-017-0879-228710743 [Google Scholar] [CrossRef] [PubMed]
[28]. Mohammad Tabar M, Mirkalantari S, Izadi Amoli R, Detection of ctx-M gene in ESBL-producing E. coli strains isolated from urinary tract infection in Semnan, IranElectron Physician 2016 8(7):2686-90.10.19082/268627648198 [Google Scholar] [CrossRef] [PubMed]
[29]. Bertrand S, Weill FX, Cloeckaert A, Vrints M, Mairiaux E, Praud K, Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003)J Clin Microbiol 2006 44:2897-903.10.1128/JCM.02549-0516891509 [Google Scholar] [CrossRef] [PubMed]
[30]. Poirel L, Decousser JW, Nordmann P, Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase geneAntimicrob Agents Chemother 2003 47:2938-45.10.1128/AAC.47.9.2938-2945.2003 [Google Scholar] [CrossRef]
[31]. Mirzaee M, Owlia P, Mansouri S, Distribution of CTX-M beta-lactamase Genes Among E. coli Strains Isolated from Patients in IranLab medicine 2009 40:724-27.10.1309/LMUUWBHMZEDYTBW5 [Google Scholar] [CrossRef]
[32]. Najar Peerayeh S, Eslami M, Memariani M, Davar Siadat S, High Prevalence of blaCTX-M-1 Group Extended-Spectrum β-lactamase Genes in E. coli Isolates From TehranJundishapur Journal of Microbiology 2013 6:e686310.5812/jjm.6863 [Google Scholar] [CrossRef]
[33]. Zong ZY, Partridge SR, Thomas L, Iredell JR, Dominance of bla(CTX-M) within an Australian extended-spectrum beta-lactamase gene poolAntimicrob Agents Chemother 2008 52:4198-202.10.1128/AAC.00107-0818725449 [Google Scholar] [CrossRef] [PubMed]
[34]. Calbo E, Freixas N, Xercavins M, Riera M, Nicolas C, Monistrol O, Foodborne nosocomial outbreak of SHV1 and CTX-M-15-producing Klebsiella pneumoniae: epidemiology and controlClin Infect Dis 2011 52:743-49.10.1093/cid/ciq23821367727 [Google Scholar] [CrossRef] [PubMed]
[35]. Malini AB, Sageerabanoo S, Kowsalya R, Gautam S, The Occurrence of CTX-M3 type extended spectrum beta lactamases among E. coli causing urinary tract infections in a tertiary care hospital in PuducherryJCDR 2012 6:1203-06. [Google Scholar]
[36]. Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strainsJ Antimicrob Chemother 2009 64:274-77.10.1093/jac/dkp19419474064 [Google Scholar] [CrossRef] [PubMed]
[37]. Korzeniewska E, Korzeniewska A, Harnisz M, Antibiotic resistant E. coli in hospital and municipal sewage and their emission to the environmentEcotoxicol Environ Saf 2013 91:96-102.10.1016/j.ecoenv.2013.01.01423433837 [Google Scholar] [CrossRef] [PubMed]
[38]. Mshana SE, Imirzalioglu C, Hossain H, Hain T, Domann E, Chakraborty T, Conjugative IncFI plasmids carrying CTX-M-15 among E. coli ESBL producing isolates at a University hospital in GermanyBMC Infect Dis 2009 9:1-8.10.1186/1471-2334-9-9719534775 [Google Scholar] [CrossRef] [PubMed]