Introduction
Malaria, a tropical disease, caused by Plasmodium is curable; still, the burden of malaria infection exists in third world countries. India, particularly among the South-East Asian region, has the maximum mortality rate. The parasites have developed resistance against antimalarial drugs. Alternatively, plants are the most important and ancient source of the drug and spices, in particular, are being used to treat various ailments.
Aim
To identify the effect of common Indian spices in targeting malarial proteases.
Materials and Methods
A bioinformatics approach was used to target malarial proteases which exhibit an important role in the erythrocytic cycle of the pathogen by degrading Haemoglobin. Proteases, in particular, Falcipain and Plasmepsin were used to perform docking studies to identify potent molecule for inhibition of malarial progression. Data were collected in the form of structural files from PDB (for protein) and PubChem (for ligands). The Protein structures and ligands were prepared and molecular docking had been done to predict interactions.
Results
All the eighteen compounds used in study have shown quite a good affinity with target proteins in the terms of energy (negative binding energy). However, comparatively, interactions of falcipain 2 with thymol and gingerol were almost three times lower than other ligand. Except these, energy ranges were in between 30-40 kilo Joule in negative. Strongest interactions were found in between Plasmepsin 4-piperamide and plasmepsin 2-gingerol. Also, it was found that gingerol was most potent bioactive molecule interacting with almost all proteins as predicted by docking studies.
Conclusion
Plasmepsin 2 and Plasmepsin 4 with gingerol and piperamide with highest cdocker energy might indicate the potential of molecules in targeting these proteases. Also, Gingerol was found to be most potent in interacting with malarial proteases.
Bioinformatics, Docking, Falcipain, Herbal
Introduction
Plants are one of the most important and diverse sources of compounds. Indian culinary spices are known to have various pharmacological activities and are treating ailments from time immortal. The World Health Organisation (WHO) estimated that approximately 65% of the world population relies on plant-derived traditional medicines for the treatment of their primary ailments. Plants are the basis of traditional medicine systems from around 2600 BC in Mesopotamia [1]. Given the paradigm shift of treatment efficacy from traditional biology to medicine, there is a need to utilise the role played by the compounds generated from the spices in various diseases [2]. In addition, molecular modelling, combinatorial chemistry, and other synthetic chemistry techniques are being used by pharmaceutical companies but particularly medicinal plants are still a huge source of new drugs, new drug leads, and New Chemical Entities (NCEs). In beginning years of the 21st decade, approximately one-quarter of the best-selling drugs worldwide were derived from natural products [2].
The vector of malarial parasite Plasmodium falciparum is the female Anopheles mosquito. The malaria parasite has a complex life cycle, leading to the challenge of developing a successful medication against it. Another problem is malarial drug resistance against existing targets [3].
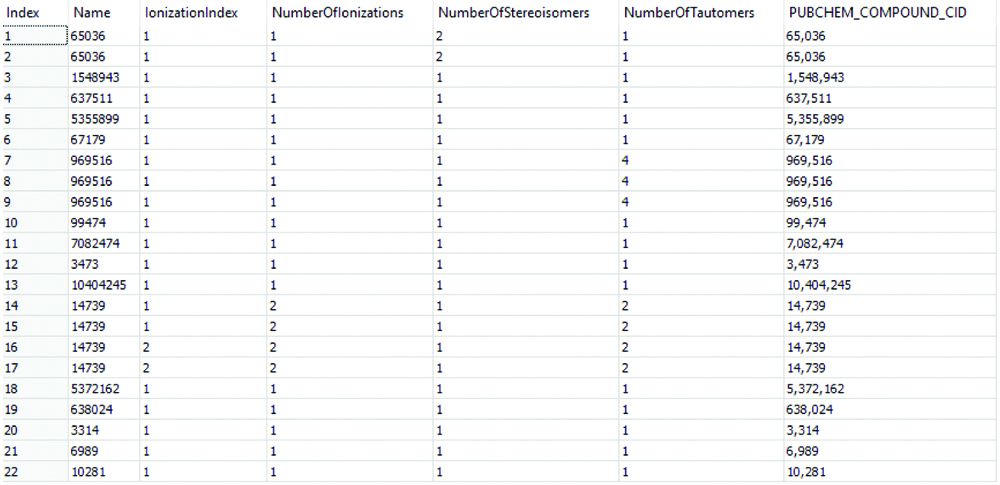
Understanding the interplay of host-parasite interaction may provide the basis for the development of a successful drug against it. The malarial life cycle consists of two cycles, an asexual cycle in the human host and a sexual cycle in the mosquito host [4].
The drug discovery for malaria was in large extent been serendipitous. Available antimalarial targets are mainly blocking blood-stage of parasites. The limitations of antimalarial drugs demand the need for new drugs, which should be ideally directed against new targets as age-old targets like are still being used [5].
Among the potential targets for antimalarial therapy are Plasmodium proteases that are drug-able targets, are being used for treating various disorders. Proteases are known to be required for the rupture and subsequent re-invasion of erythrocytes and for the degradation of haemoglobin [6,7]. Because of the high degree of similarity, the specific role of the food vacuole plasmepsins in vivo has been unclear [8-10]. This study was focused to discover a lead molecule from spices which can be used to target malarial proteases. Falcipain and plasmepsins were being used in this study. Molecular docking had been used to predict the interactions between proteins and bioactive molecules. The effect of some spices was established to have anti-malarial activity. Janfaza S and Janfaza E, illustrated that the antimalarial activity of Thymoquinone from oil of Nigella sativa has antimalarial properties [11] and also thymoquinone and artemisinin hybrid molecules had shown to have increased potency as tested in vitro [12]. Antimalarial activity of herbal extract and formulations from nutmeg had been proved in vitro [13]. Diosgenone (a compound derived from diosgenin) derivatives reduced Malarial parasitemia in mice by oral administration [14]. Chakrabarti R et al., illustrated inhibitory effect of curcumin [15]. Reddy R et al., showed that curcumin inhibits chloroquine-resistant Plasmodium falciparum growth in culture and oral administration in infected mice reduces blood parasitemia by 80-90 percent [16]. Curcumin artemisinin combination therapy can also be used for treating malaria as suggested in vitro [17]. Cinnamic acid derivatives were also reported to inhibit in vitro growth of Plasmodium falciparum [18]. Allicin could partially protect host against malaria parasite [19] and also shown to inhibit circumsporozoite protein of Plasmodium [20].
In a country like India, where using plant parts as medicine being a tradition and still most of the population relies on traditional medicine, use of spice derived lead compounds can pave a way for better treatment of malaria. So the present work was aimed to predict the potential lead from spices namely {(Galbanic Acid (Asafetida), Eugenol (Basil and clove), Allicin (garlic), Curcumin (Turmeric), Thymol (Carom seeds), Thymoquinone (Black Cumin), Anethole (Fennel), Diosgenin (Fenugreek), Macelignan (Nutmeg), Piperamide (Black pepper), Pipercide (Black pepper), Capsaicin (Chillies), Gingerol (Ginger), Cinnamic acid (Cinnamon), Cinnamaldehyde (Cinnamon)} that can target malarial Proteases and can provide a better treatment for malaria.
Materials and Methods
The current study was completely an In silico analysis. It was conducted in the year 2017. No ethical approval was involved as complete study is based on bioinformatics. The study was conducted between February to October 2017 at Birla Institute of Scientific Research, Jaipur, Rajasthan, India as Doctoral research from KIIT University, Bhubaneswar, Odisha, India.
Retrieval of Structural Files of Bio-active Compounds of Spices
The bio-active compounds of spices were known to have medicinal properties [21-25]. These compounds were archived in their three-dimensional structure. PubChem (https://pubchem.ncbi.nlm.nih.gov/) was used to retrieve structural files in SDF format of these compounds [26]. The study was aimed to find out the effect of common culinary spices used in India against malarial proteases.
Retrieval of Protein Structural Data
RCSB Protein Data Bank (PDB) is one of the major sources of Protein structural data [27]. Structure of Malarial proteins has been retrieved from PDB database. The details of structures taken had been described in [Table/Fig-1] [28-32].
Proteins used for docking Studies [28-32].
| Sr. No. | Protein name | PDB ID | Resolution | Active site origin | Active site | Reference |
|---|
| 1. | Falcipain 3 | 3BWK | 2.42 Ao | PDB | -6.66929, -37.5781, 49.00 R=10.7 | [28] |
| 2. | Falcipain 2 | 3PNR | 2.60 Ao | PDB | 14.9119, 12.5747, 10.3243 R=17.1 | [29] |
| 3. | Plasmepsin 1 | 3QRV | 2.40 Ao | Receptor cavity | 14.4615, 18.618, 24.092 R=24.3 | [30] |
| 4. | Plasmepsin 2 | 1XDH | 1.70 Ao | PDB | 16.3857, 62.7968, 24.2156 R=11.5 | [31] |
| 5. | Plasmepsin 4 | 1LS5 | 2.80 Ao | PDB | -26.4472, 36.8228, 43.1958 R=11.7 | [32] |
Preparation of Protein Structure
Discovery studio (Version) of Biovia systems (Version 2017 R2) was used for preparing Protein structure. The licensed modules were used for performing all the analysis [33].
Active Site Prediction
The prediction of active site was done using Discovery studio. Selection was made using active sites present in PDB record. However, Active sites can also be selected using Protein cavities [Table/Fig-2].
List of Bio-actives and corresponding spices.
| Active compound (Source spice) | Pub chem ID | Molecular formula and weight (g/moL) | Molecular structure |
|---|
| Galbanic acid (Asafoetida) | 7082474 | C24H30O5 (398.499) | 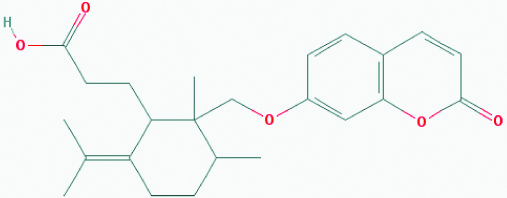 |
| Eugenol (Clove/Basil) | 3314 | C10H12O2 (164.204) | 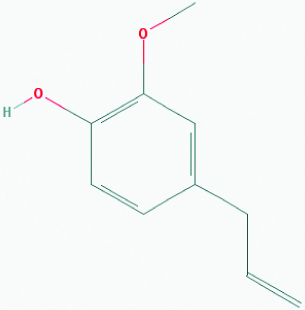 |
| Thymoquinone (Black cumin) | 10281 | C10H12O2 (164.204) | 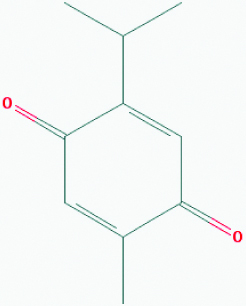 |
| Thymol (Carom seeds) | 6989 | C10H14O (150.221) | 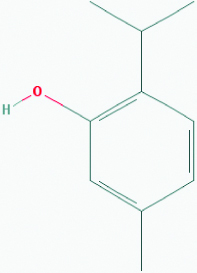 |
| Macelignan (Nutmeg) | 10404245 | C20H24O4 (328.408) | 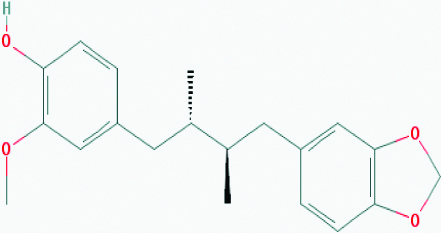 |
| Trans-Anethole (Fennel) | 637563 | C10H12O (148.205) | 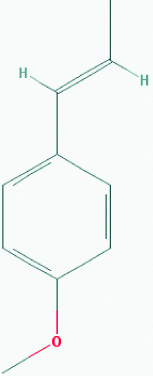 |
| Diosgenin (Fenugreek) | 99474 | C27H42O3 (414.63) | 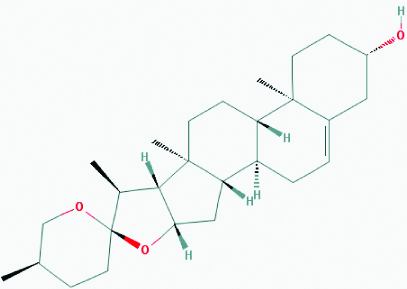 |
| Piperamide (Black pepper) | 14739 | C17H28N4O (304.438) | 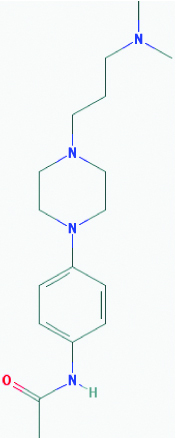 |
| Piperine (Black pepper) | 638024 | C17H19NO3 (285.343) | 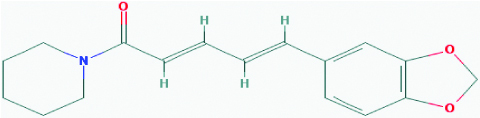 |
| Retrofractamide B (Pipercide) (Black pepper) | 5372162 | C22H29NO3 (355.478) | 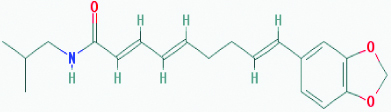 |
| Capsaicin (Chillies) | 1548943 | C18H27NO3 (305.418) | 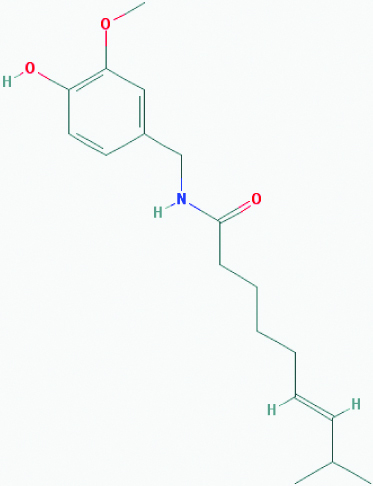 |
| Coriandrol (Coriander) | 67179 | C10H18O (154.253) | 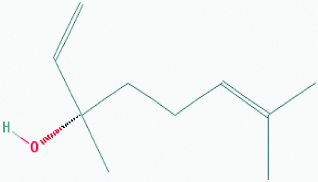 |
| Ursolic acid (Basil) | 64945 | C30H48O3 (456.711) | 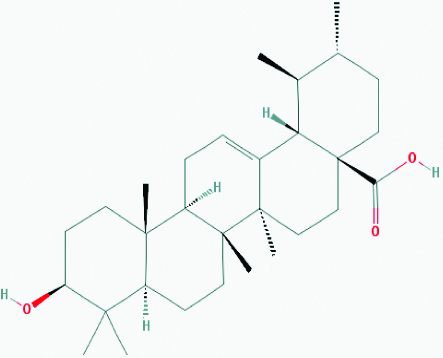 |
| Cinnamaldehyde (Cinnamon) | 637511 | C9H8O (132.162) | 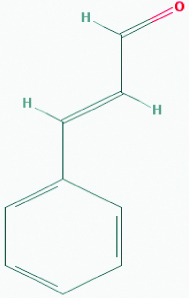 |
| Cinnamic acid (Cinnamon) | 444539 | C9H8O2 (148.161) | 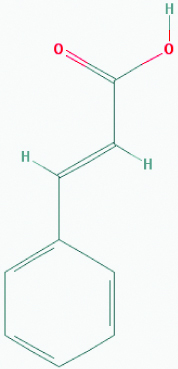 |
| Gingerol (Ginger) | 3473 | C17H26O4 (294.391) | 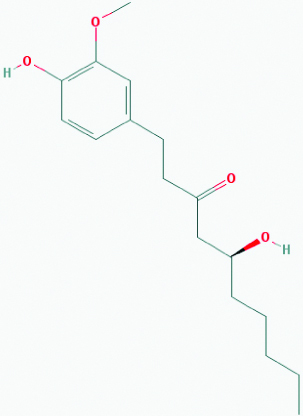 |
| Allicin (Garlic) | 65036 | C6H10OS2 (162.265) | 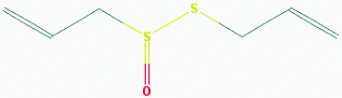 |
| Curcumin (Turmeric) | 969516 | C21H20O6 (368.385) | 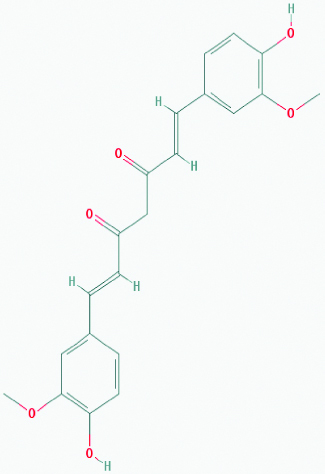 |
Ligand Preparation for Docking
The ligands were prepared using Discovery Studio produced stereoisomers; tautomer checked charges and ligand/ligands were ready for docking. Already known bio-active molecules from different spices were retrieved from the PubChem Database in SDF format and merged into same Spatial Data File (SDF) file. The molecules were then prepared using Ligand preparation tool of Discovery studio.
Molecular Docking Studies
Molecular docking was used to screen compounds from the library of prepared ligands which were showing a reasonable interaction with proteins of interest. Docker method was used for docking. Docker energy was one of the important factors in screening of ligand poses besides the interaction type and the bond length.
Results
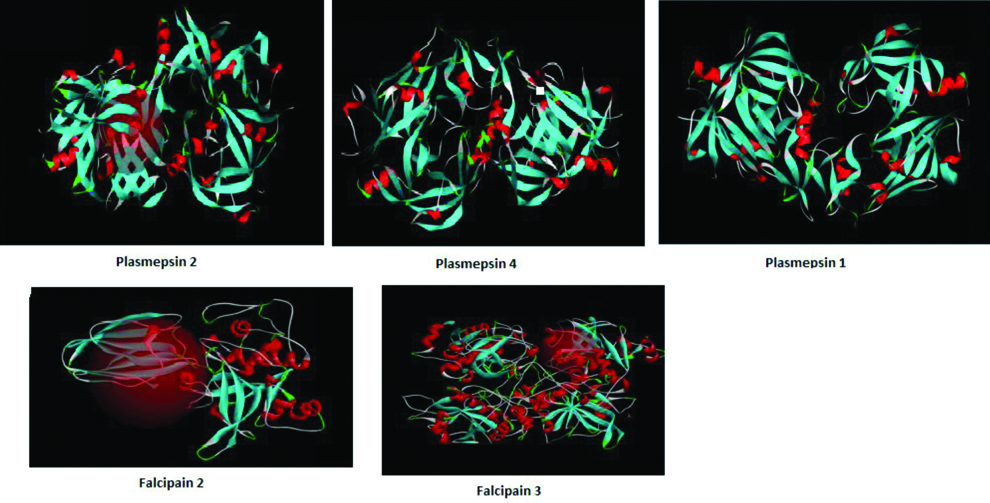
Six Structures were Prepared in silico
The PDB structures of six proteins, three falcipains and three plasmepsins were analysed using Biovia discovery studio tool prepare protein.
The Prepare Protein protocol prepared proteins for input into other protocols, performing tasks such as inserting missing atoms in incomplete residues, modelling missing loop regions, deleting alternate conformations (disorder), removing waters, standardising atom names and protonating titratable residues using predicted pKs [Table/Fig-3].
Structures of plasmepsins and falcipains.
Ligand Analysis and Preparation
A total of 18 compounds were taken for docking studies and ligand preparation tool is used to prepare 18 ligands for docking. The Prepare ligands protocol helped to prepare ligands for input into other protocols, performing tasks such as removing duplicates, enumerating isomers and tautomers, and generating 3D conformations. A total of 22 ligands were generated including tautomers and stereoisomers [Table/Fig-4]. Allicin (PID 65036) had 2 stereoisomers, Curcumin (969516) had 4 tautomers, piperamide had two ionizations and two tautomers for each. A ligand was failed due to duplicate result and that was one of the structures of 969516. So, we had a total of 22 structures to be docked against 5 proteins [Table/Fig-4].
Ligands used for docking.
Discussion
Current study was based on semi flexible docking scheme in which protein remains rigid and ligand tries to fit into the active pocket. All the five proteins had been screened against 22 prepared ligands which yielded hundreds of ligand poses [Table/Fig-5,6]. Initially, the ligand with highest Cdocker energy for all proteins was selected. Hydrogen bonds and hydrophobic interaction play an important role in protein ligand interaction. Presence of H bond indicates a strong interaction between protein and ligand [34-36]. The ligand poses with higher Cdocker energy aligns well with the structure. Lowest energy (Highest negative) indicates a more favourable binding of ligand [37-39].
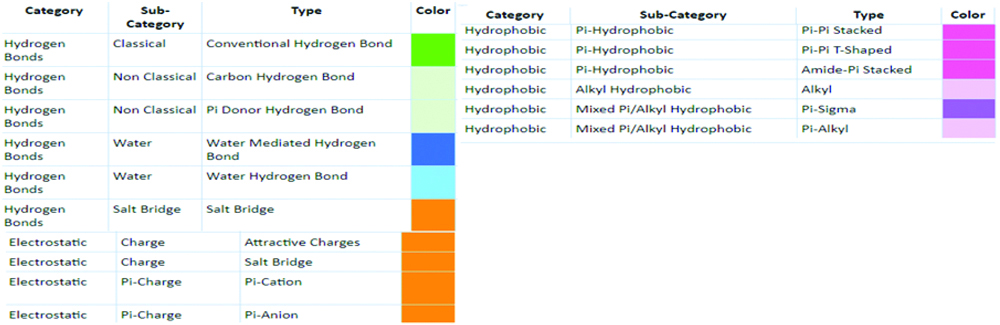
Color coding scheme for ligand interaction.
A 2D Interaction diagram of top energy poses.
| Protein (C Docker energy) | Interacting molecule | 2D Interaction map |
|---|
| Plasmepsin 4 (40.8051) | Piperamide | 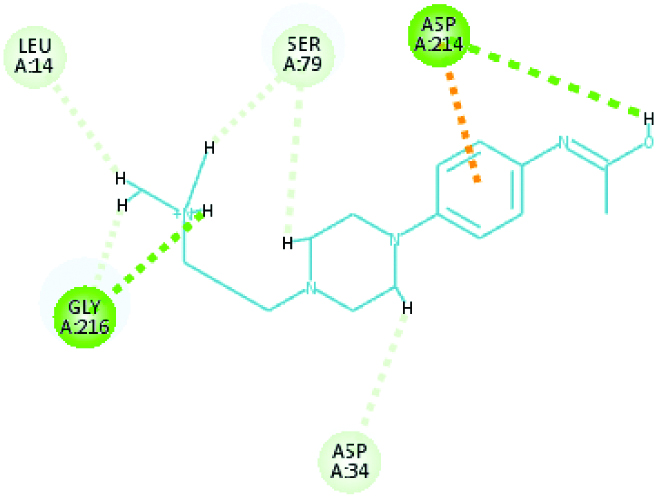 |
| Plasmepsin 4 (39.5709) | Gingerol | 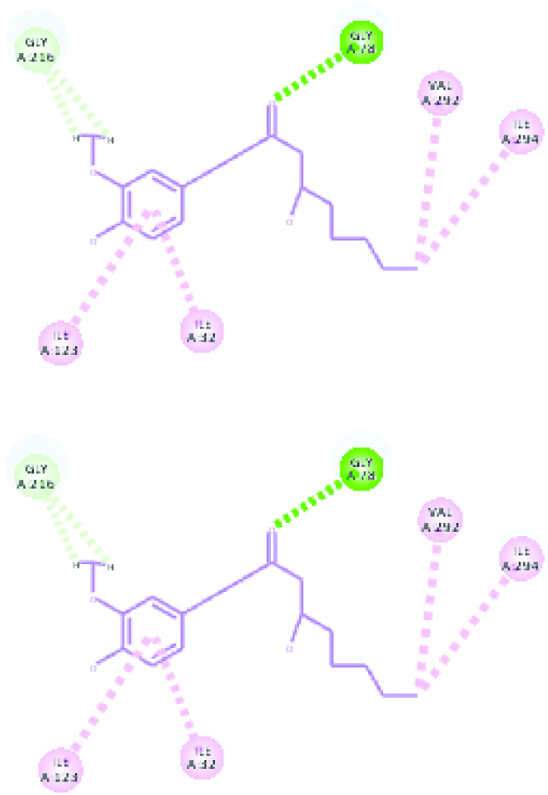 |
| Plasmepsin 2 (40.4649) | Gingerol | 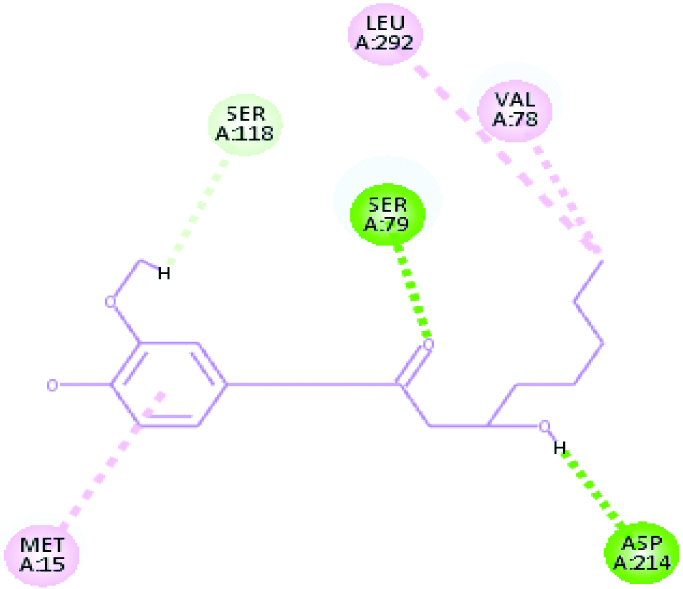 |
| Falcipain 3 (27.7347) | Capsaicin | 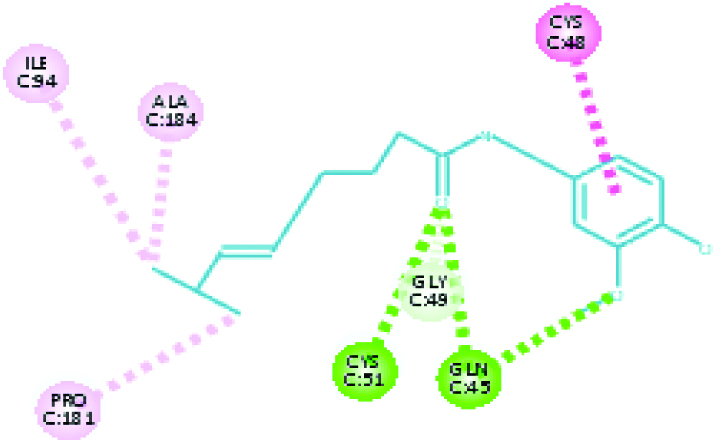 |
| Falcipain 3 (32.3169) | Gingerol | 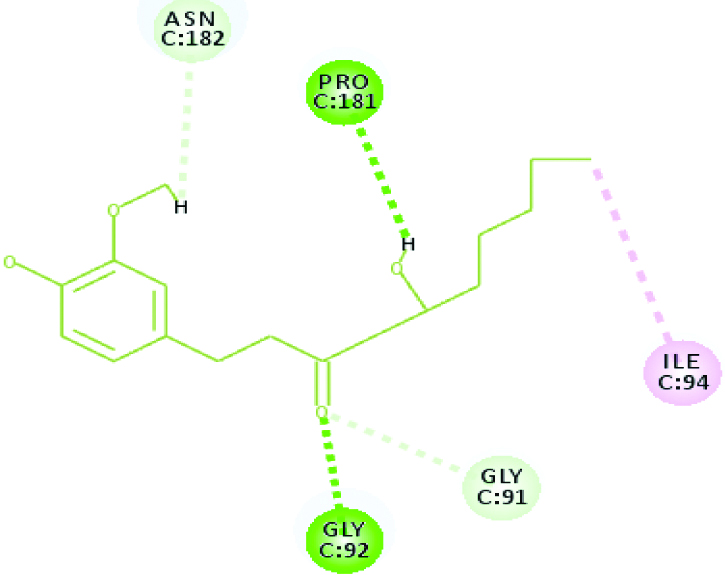 |
| Falcipain 2 (10.9035) | Thymol | 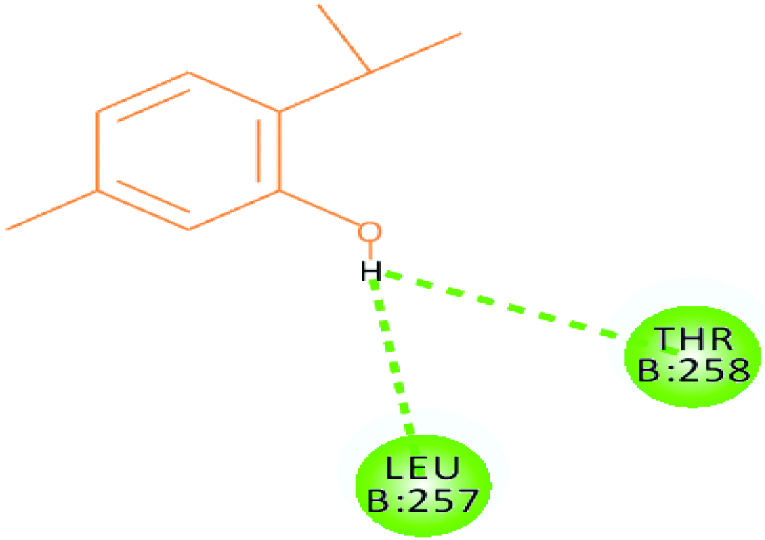 |
| Falcipain 2 (14.2815) | Gingerol | 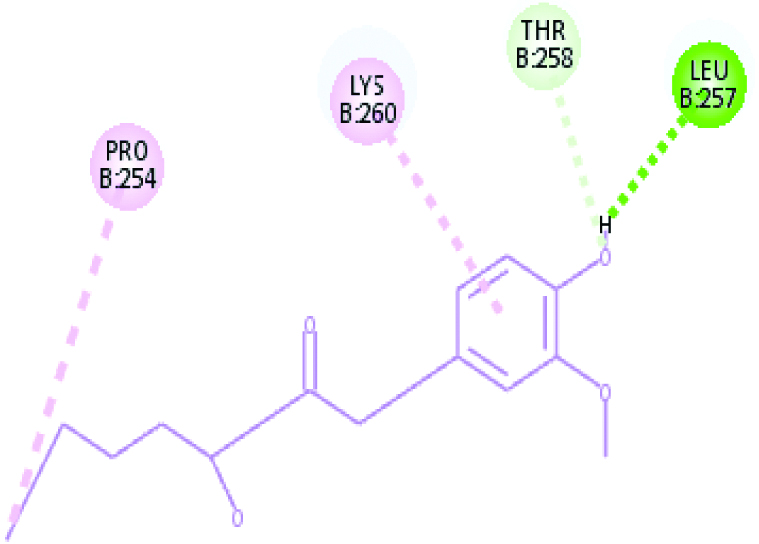 |
| Plasmepsin I (32.096) | Gingerol | 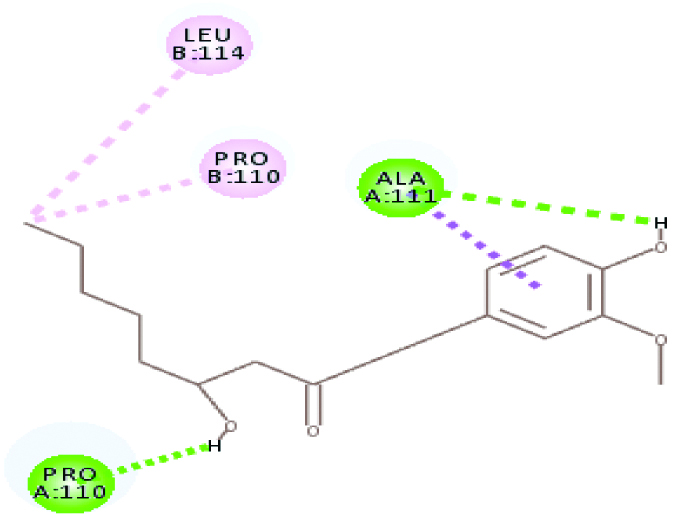 |
| Plasmepsin I (31.6361) | Piperamide | 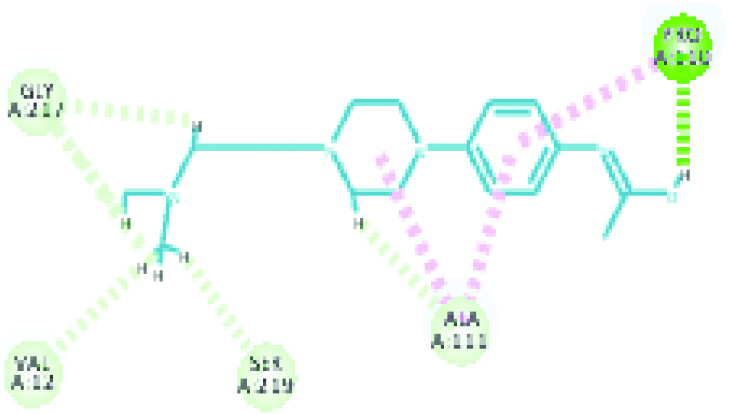 |
| Plasmepsin I (30.5079) | Curcumin | 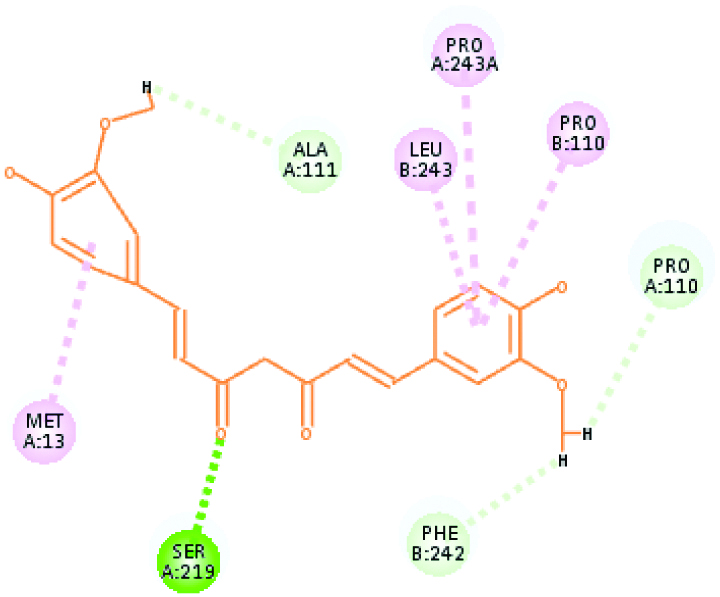 |
These two criteria were considered here for selecting poses. Hence, interactions of plasmepsin 4-piperamide [Table/Fig-7] and plasmepsin 2-gingerol [Table/Fig-8] simultaneously had highest Cdocker energy. The polar amino acids i.e., Serine and aspartate present at active site of plasmepsins seem to have important role in interaction.
Plasmepsin-Piperamide interaction and Plasmepsin structure.
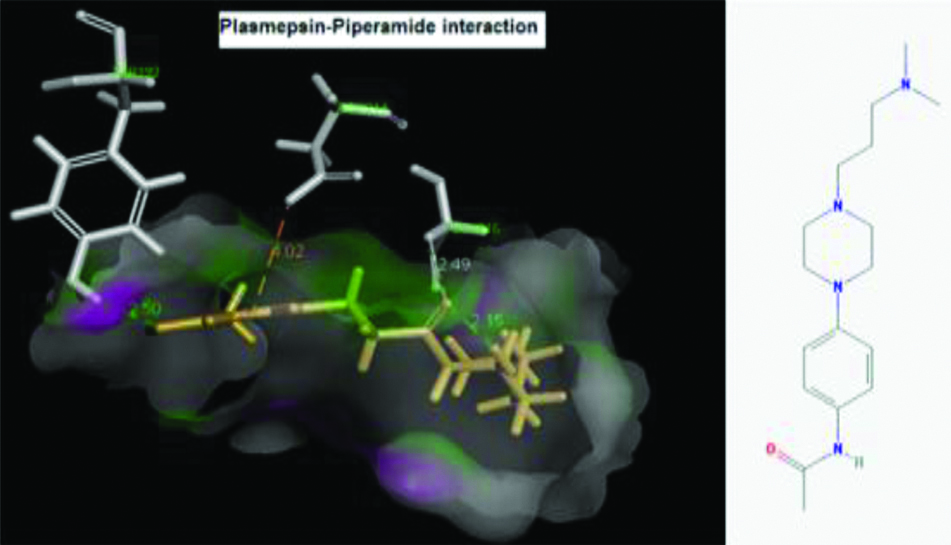
Plasmepsin-Gingerol interaction and Gingerol structure.
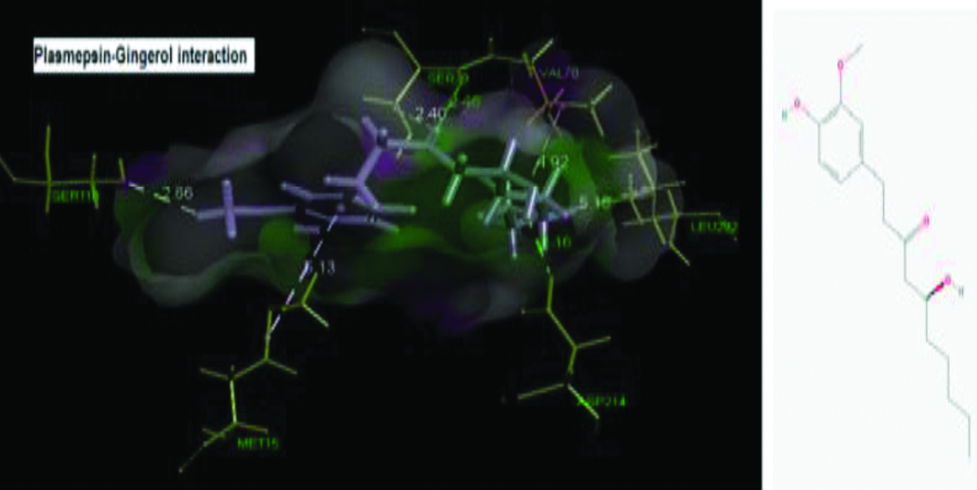
However, if we assume the scenario in context of ligands, only few ligands had shown interaction with malarial proteases, most potent being gingerol, interacting with most of the structure of malarial proteases.
Conclusion
The docking based virtual screening was done for 22 prepared ligands and for five protease structure (two falcipain and three plasmepsins). Plasmepsin 2 and Plasmepsin 4 with gingerol and piperamide with highest cdocker energy might indicate the potential of molecules in targeting these proteases. Also, gingerol was found to be most potent in interacting with malarial proteases further modification and development of analogues for this ligand may lead to development of better antimalarial drug.
[1]. Cragg GM, Newman DJ, Natural Products: A continuing source of novel drug leadsBiochem Biophys Acta 2013 1830(6):3670-95.10.1016/j.bbagen.2013.02.00823428572 [Google Scholar] [CrossRef] [PubMed]
[2]. Balunas M, Kinghorn A, Drug discovery from medicinal plantsLife Sciences 2005 78(5):431-41.10.1016/j.lfs.2005.09.01216198377 [Google Scholar] [CrossRef] [PubMed]
[3]. Silva A, Lee A, Gulnik S, Maier P, Collins J, Bhat T, Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparumProceedings of the National Academy of Sciences 1996 93(19):10034-39.10.1073/pnas.93.19.100348816746 [Google Scholar] [CrossRef] [PubMed]
[4]. Teixeira C, Gomes JR, Gomes P, Falcipains, plasmodium falciparum cysteine proteases as key drug targets against malariaCurrent Medicinal Chemistry 2011 2012:34519510.2174/092986711795328328 [Google Scholar] [CrossRef]
[5]. Li H, Child M, Bogyo M, Proteases as regulators of pathogenesis: Examples from the ApicomplexaBiochimicaet Biophysica Acta (BBA)-Proteins and Proteomics 2012 1824(1):177-85.10.1016/j.bbapap.2011.06.00221683169 [Google Scholar] [CrossRef] [PubMed]
[6]. Pandey KC, Dixit R, Structure-function of falcipains: malarial cysteine proteasesJournal of Tropical Medicine 2012 :1-11.10.1155/2012/34519522529862 [Google Scholar] [CrossRef] [PubMed]
[7]. Rosenthal P, Proteases of malaria parasites: new targets for chemotherapyEmerging Infectious Diseases 1998 4(1):49-57.10.3201/eid0401.9801079452398 [Google Scholar] [CrossRef] [PubMed]
[8]. Liu J, Gluzman I, Drew M, Goldberg D, The Role of Plasmodium falciparum food vacuole plasmepsinsJournal of Biological Chemistry 2004 280(2):1432-37.10.1074/jbc.M40974020015513918 [Google Scholar] [CrossRef] [PubMed]
[9]. Moura P, Dame J, Fidock D, Role of plasmodium falciparum digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitorsAntimicrobial Agents and Chemotherapy 2009 53(12):4968-78.10.1128/AAC.00882-0919752273 [Google Scholar] [CrossRef] [PubMed]
[10]. Berry C, New targets for antimalarial therapy: The plasmepsins, malaria parasite aspartic proteinasesBiochemical Education 1997 25(4):191-94.10.1016/S0307-4412(97)00130-1 [Google Scholar] [CrossRef]
[11]. Janfaza S, Janfaza E, The study of pharmacologic and medicinal valuation of thymoquinone of oil of Nigella sativa in the treatment of diseasesScholars Research Library 2012 3:1953-57. [Google Scholar]
[12]. Fröhlich T, Reiter C, Saeed M, Hutterer C, Hahn F, Leidenberger M, Friedrich O, Synthesis of thymoquinone-artemisininin hybrids: new potent antileukemia, antiviral, and antimalarial agentsACS Med Chem Lett 2018 9(6):534-39.10.1021/acsmedchemlett.7b0041229937978 [Google Scholar] [CrossRef] [PubMed]
[13]. Thiengsusuk A, Chaijaroenkul W, Na-Bangchang K, Antimalarial activities of medicinal plants and herbal formulations used in Thai traditional medicineParasitology Research 2013 112(4):1475-81.10.1007/s00436-013-3294-623340720 [Google Scholar] [CrossRef] [PubMed]
[14]. Pabón A, Escobar G, Vargas E, Cruz V, Notario R, Blai S, Diosgenone synthesis, anti-malarial activity and qsar of analogues of this natural productMolecules 2013 18(3):3356-78.10.3390/molecules1803335623493102 [Google Scholar] [CrossRef] [PubMed]
[15]. Chakrabarti R, Rawat P, Cooke B, Coppel R, Patankar S, Cellular effects of curcumin on plasmodium falciparum include disruption of microtubulesPLoS ONE 2013 8(3):e5730210.1371/journal.pone.005730223505424 [Google Scholar] [CrossRef] [PubMed]
[16]. Reddy R, Vatsala P, Keshamouni V, Padmanaban G, Rangarajan P, Curcumin for malaria therapyBiochemical and Biophysical Research Communications 2005 326(2):472-74.10.1016/j.bbrc.2004.11.05115582601 [Google Scholar] [CrossRef] [PubMed]
[17]. Nandakumar D, Nagaraj V, Vathsala P, Rangaraja P, Padmanaba G, Curcumin-artemisininin combination therapy for malariaAntimicrobial Agents and Chemotherapy 2006 50(5):1859-60.10.1128/AAC.50.5.1859-1860.200616641461 [Google Scholar] [CrossRef] [PubMed]
[18]. Kanaani J, Ginsburg H, Effects of cinnamic acid derivatives on in vitro growth of Plasmodium falciparum and on the permeability of the membrane of malaria-infected erythrocytesAntimicrobial Agents and Chemotherapy 1992 36(5):1102-08.10.1128/AAC.36.5.11021510401 [Google Scholar] [CrossRef] [PubMed]
[19]. Feng Y, Zhu X, Wang Q, Jiang Y, Shang H, Cui L, Allicin enhances host pro-inflammatory immune responses and protects against acute murine malaria infectionMalaria Journal 2013 11(1):26810.1186/1475-2875-11-26822873687 [Google Scholar] [CrossRef] [PubMed]
[20]. Coppi A, Cabinian M, Mirelman D, Sinnis P, Antimalarial activity of allicin, a biologically active compound from garlic clovesAntimicrobial Agents and Chemotherapy 2006 50(5):1731-37.10.1128/AAC.50.5.1731-1737.200616641443 [Google Scholar] [CrossRef] [PubMed]
[21]. Ali B, Blunden G, Tanira M, Nemmar A, Some phytochemical, pharmacological and toxicological properties of ginger (Zingiberofficinale Roscoe): A review of recent researchFood and Chemical Toxicology 2008 46(2):409-20.10.1016/j.fct.2007.09.08517950516 [Google Scholar] [CrossRef] [PubMed]
[22]. Amalraj A, Pius A, Gopi S, Gopi S, Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives-A reviewJournal of Traditional and Complementary Medicine 2017 7(2):205-33.10.1016/j.jtcme.2016.05.00528417091 [Google Scholar] [CrossRef] [PubMed]
[23]. Gupta M, Pharmacological properties and traditional therapeutic uses of important Indian spices: a reviewInternational Journal of Food Properties 2010 13(5):1092-116.10.1080/10942910902963271 [Google Scholar] [CrossRef]
[24]. Parthasarathy V, Chempakam B, Zachariah T, Chemistry of spices 2008 Wallingford, UKCABI Pub10.1079/9781845934057.0000 [Google Scholar] [CrossRef]
[25]. Opara E, Chohan M, Culinary herbs and spices: their bioactive properties, the contribution of polyphenols and the challenges in deducing their true health benefitsInternational Journal of Molecular Sciences 2014 15(12):19183-202.10.3390/ijms15101918325340982 [Google Scholar] [CrossRef] [PubMed]
[26]. Fu G, Batchelor C, Dumontier M, Hastings J, Willighagen E, Bolton E, PubChem RDF: towards the semantic annotation of PubChem compound and substance databasesJournal of Chem Informatics 2015 7:3410.1186/s13321-015-0084-426175801 [Google Scholar] [CrossRef] [PubMed]
[27]. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, The protein data bankNucleic Acids Research 2000 28:235-42.10.1093/nar/28.1.23510592235 [Google Scholar] [CrossRef] [PubMed]
[28]. Kerr ID, Lee JH, Farady CJ, Marion R, Rickert M, Sajid M, Vinyl sulfones as antiparasitic agents and a structural basis for drug designJ Biol Chem 2009 284:25697-703.10.1074/jbc.M109.01434019620707 [Google Scholar] [CrossRef] [PubMed]
[29]. Hansen G, Heitmann A, Witt T, Li H, Jiang H, Shen X, Structural basis for the regulation of cysteine-protease activity by a new class of protease inhibitors in PlasmodiumStructure 2011 19:919-29.10.1016/j.str.2011.03.02521742259 [Google Scholar] [CrossRef] [PubMed]
[30]. Bhaumik P, Horimoto Y, Xiao H, Miura T, Hidaka K, Kiso Y, Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparumJ Struct Biol 2011 175:73-84.10.1016/j.jsb.2011.04.00921521654 [Google Scholar] [CrossRef] [PubMed]
[31]. Prade L, Structure of plasmepsin II in complex with pepstatin analougeProtein Data Bank[Available form https://www.rcsb.org/structure/1XE6] [Google Scholar]
[32]. Asojo OA, Gulnik SV, Afonina E, Yu B, Ellman JA, Haque TS, Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparumJ Mol Biol 2003 327:173-81.10.1016/S0022-2836(03)00036-6 [Google Scholar] [CrossRef]
[33]. Discovery Studio Predictive Science Application | Dassault Systèmes BIOVIA [Internet]. Accelrys.com. 2018 Available from: http://accelrys.com/products/collaborative-science/biovia-discovery-studio/ [Google Scholar]
[34]. Goodford P, Structure and function of water in drug-receptor interactionsJournal of Molecular Graphics 1990 8(4):233-34.10.1016/0263-7855(90)80023-9 [Google Scholar] [CrossRef]
[35]. Patil R, Das S, Stanley A, Yadav L, Sudhakar A, Varma A, Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designingPLoS ONE 2010 5(8):e1202910.1371/journal.pone.001202920808434 [Google Scholar] [CrossRef] [PubMed]
[36]. Musyoka T, Kanzi A, Lobb K, Tastan Bishop Ö, Structure based docking and molecular dynamic studies of plasmodial cysteine proteases against a South African natural compound and its analogsScientific Reports 2016 6:2369010.1038/srep2369027030511 [Google Scholar] [CrossRef] [PubMed]
[37]. Azam S, Abbasi S, Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routinesTheoretical Biology and Medical Modelling 2013 10(1):6310.1186/1742-4682-10-6324156411 [Google Scholar] [CrossRef] [PubMed]
[38]. El-Sayed M, Abdel-Aziz N, Abdel-Aziz A, El-Azab A, Asiri Y, ElTahir K, Design, synthesis, and biological evaluation of substituted hydrazone and pyrazole derivatives as selective COX-2 inhibitors: Molecular docking studyBioorganic & Medicinal Chemistry 2011 19(11):3416-24.10.1016/j.bmc.2011.04.02721570309 [Google Scholar] [CrossRef] [PubMed]
[39]. Singh S, Srivastava P, Molecular docking studies of myricetin and its analogues against human PDK-1 kinase as candidate drugs for cancerComputational Molecular Bioscience 2015 5:20-33.10.4236/cmb.2015.52004 [Google Scholar] [CrossRef]