Nasal carriage of MRSA by apparently healthy populations represents a major risk factor for subsequent infection and its transmission to vulnerable individuals with weakened immune systems in a community. Hence, this study determined the antibiogram and virulent characteristics of Staphylococcus aureus isolated from the nasal cavity of healthy students of Niger Delta University, Amassoma, Bayelsa State, Nigeria as means of adopting effective measures for the treatment and prevention of its infections transmission to the University community and the neighbouring environments.
Materials and Methods
Study Population
The apparently healthy undergraduate students of College of Health Sciences in Niger Delta University, Amassoma, Bayelsa State, who had not taken a course of antimicrobial therapy nor admitted in a hospital within a period of three months prior to the commencement of the survey were recruited into this study. Niger Delta University is one of the fast growing State Universities in Nigeria, with its location in the Southern part of Nigeria and majority of its students are from the Southern region of the country. The volunteers in this cross-sectional study were randomly selected from the University lecture rooms and they gave written informed consent before the collection of nasal specimens. The basic demographic data such as age and sex (excluding names) were obtained from the volunteers and the study was approved by the ethical committee of the institution before the commencement of the study which spanned through April to October 2016. The sample size for this study was determined using Cochran’s formula as described by Bartlett JE et al., [13].
Collection of Samples and Bacteriology
Nasal specimens of 400 healthy volunteers were collected with sterile swab sticks moistened with sterile normal saline solution (0.9%w/v Sodium chloride) and transferred immediately to the Department of Pharmaceutical Microbiology and Biotechnology laboratory for inoculation and streaking on the already prepared sterilised Nutrient Agar plates. The discrete colonies obtained from the inoculated Nutrient agar (Oxoids, UK) plates after incubation at 37°C for 24 hour were sub-cultured on Mannitol Salt Agar (Oxoids, UK) plates and then incubated at 35°C for 24-48 hour. The characteristic colonies on the Mannitol Salt agar plates were identified using standard established microbiological methods, which include colonial morphology, Gram’s stain reaction and biochemical characteristics. The isolates that were Gram-positive grape-like clustered cocci, positive to catalase test, mannitol fermentation test, DNase test and slide coagulase test were subjected to PCR analysis for the presence of nuc gene as a confirmation for S. aureus [14,15].
Extraction of DNA from Biochemically Confirmed S. aureus
A single colony of each of the biochemically confirmed S. aureus isolates was cultured in sterilised Luria-Bertani broth (Oxoids) and the bacterial cells were recovered after centrifugation. The cells were emulsified in 0.5 ml of sterile DNase-free biological grade water (Inqaba Biotec), boiled at 100°C for 10 minutes in a thermal block and rapidly cooled in a freezer for 30 minutes. The mixture was centrifuged at 10,000 rpm for 10 minutes, and supernatant containing DNA collected, quantified using Nano drop 1000 and stored at 4°C until further investigation [16].
Screening for S. aureusnuc and mecA Genes by PCR Amplification
PCR was used to separately amplify S. aureus specific nuc gene and methicillin resistant mecA gene using the primers described by Ali R et al., [15] and Nakagawa S et al., [17]. The S. aureus specific nuc gene was amplified using a 21-nucleotide forward primer 5’-GCGATTGATGGTGATACGGTT-3’ and 25-nucleotide reverse primer 5’-AGCCAAGCCTTGAACGAACTAAAGC-3’ with a product size of 270 bp while the mecA gene was amplified using a 20-nucleotide of both primers forward 5’-CGGTAACATTGATCGCAACG-3’and reverse 5’-TTTGCCAACCTTTACCATCG-3’ with a product size of 154 bp. The PCR was performed in a 20 μL reaction mixture containing 1X taq master mix (Taq polymerase, dNTPs, MgCl2 buffer), the primers (forward and reverse) at a concentration of 0.4M, water and the extracted DNA as template. The nuc gene was amplified at DNA denaturation at 94°C for 5 minutes, 94°C for 60 seconds, primers annealing at 55°C for 30 seconds, extension at 72°C for 60 seconds, 72°C for 7 minutes for 37 cycles while the MecA gene was amplified at DNA denaturation at 95°C for 15 minutes, 95°C for 30 seconds, primers annealing at 55°C for 40 seconds, extension at 72°C for 50 seconds, 72°C for 20 minutes for 35 cycles. The amplified PCR products were analysed on a 2% agarose gel (Biogene, UK) stained with ethidium bromide (Sigma, UK) and electrophoresis was performed in 1X TBE buffer at 120 V for 30 minutes with a 100 bp molecular marker. The gels were subsequently visualised using UV illuminator and photographed.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of all S. aureus strains was determined for amoxicillin/clavulanic acid (AMC 20/10 μg), erythromycin (E 15 μg), co-trimoxazole (trimethoprim/sulfamethoxazole) (SXT 1.25/23.75 μg), tetracycline (TE 30 μg), gentamycin (CN 10 μg), cefoxitin (FOX 30 μg), ciprofloxacin (CIP 5 μg) and levofloxacin (LEV 5 μg) (Qxoids, UK) using the modified Kirby-Bauer disc diffusion technique in accordance with the Clinical and Laboratory Standard Institute guidelines [18]. The S. aureus isolates that were resistant to at least one agent in three or more of the eight classes of antimicrobial agent used in this study were defined as having MDR [19].
Screening for Haemolysin Production in the S. aureus Isolates
Haemolytic property of the S. aureus isolates was determined by inoculating the isolates on freshly prepared sterile 5%v/v Blood Agar (consisting 5 mL of human blood in 100 mL of Nutrient Agar) plates using straight wire loop and incubated at 37°C for 24 hours. Thereafter, plates were observed for green to dark colouration of the agar (partial lysis of red blood cells-alpha haemolysis) and clear zones (complete lysis of red blood cells-beta haemolysis) around inoculated organisms, indicating the production of haemolysin.
Screening for Biofilm Production in the S. aureus Isolates
Biofilm production in S. aureus isolates was performed using Congo Red Agar medium which was prepared using the combination of brain heart infusion agar 52 g/L, sucrose 50 g/L and Congo red indicator 8 g/L as described by Mathur T et al., [20]. The Congo red was prepared as concentrated aqueous solution separately from other medium constituents and then sterilised in different containers before adding them together when the agar had cooled to 55°C before distributing to sterile plates to solidify. The plates were then inoculated with the test organisms and incubated at 37°C for 24 hours before examining for black colonies with a dry crystalline consistency indicating biofilm production.
Statistical Analysis
The groups differences were tested using the Chi-square test (or Fisher’s-exact test when expected frequencies were too low), with the assumed level of statistical significance at a p-value of <0.05. Data analysis was performed with SPSS version 15.0 for Windows (SPSS Inc, USA).
Results
The Prevalence, Distribution and Confirmation of S. aureus Isolates
The 400 nasal specimens from females 276 (69%) and males 124 (31%) volunteers of ages 15-35 years, yielded 183 (45.8%) significant growth of Staphylococci. Only 47 (11.8%) S. aureus isolates were recovered through the biochemical tests and the PCR targeted S. aureus specific nuc gene [Table/Fig-1]. The age distribution of the volunteers with the S. aureus carriage is shown in [Table/Fig-2]. The gender distribution of the volunteers with S. aureus isolates was females 25 (9.1%; 25/276) and males 22 (17.7%; 22/124). The nasal carriage of S. aureus isolates was found to be significantly higher in the males than the females (Odds ratio=2.165; χ2=5.413; p=0.013) [Table/Fig-3].
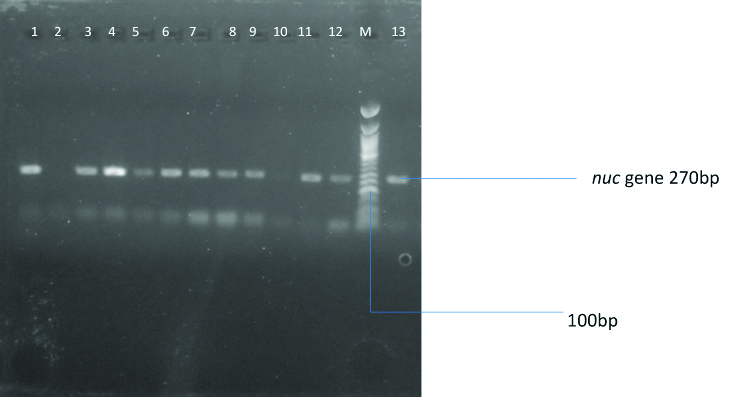
Agarose gel showing the amplified nuc gene of S. aureus isolates. Lanes 1, 3, 4, 5, 6, 7-9 and 11-13 represents the positive bands while Lane M represents the 100bp Quick-Load Molecular ladder respectively.
The age distribution of the volunteers with nasal S. aureus.
| Age (yrs) | No. of sample | S. aureus (%) |
|---|
| 15-19 | 48 | 5 (10.4) |
| 20-24 | 243 | 27 (11.1) |
| 25-30 | 84 | 12 (14.3) |
| 31-35 | 25 | 3 (12) |
| Total | 400 | 47 (11.8) |
The gender distribution of the volunteers with nasal S. aureus.
| Gender | No. of sample | Detection of S. aureus (%) | Chi-square | p-value |
|---|
| Yes | No |
|---|
| Females | 276 | 25 (9.1) | 251 (90.9) | 5.413 | 0.013* |
| Males | 124 | 22 (17.7) | 102 (82.3) |
| Total | 400 | 47 (11.8) | 353 (88.2) | | |
*Statistically significant (p<0.05)
Prevalence of Virulence among the S. aureus Isolates
A total of 37 (78.7%) S. aureus isolates phenotypically expressed haemolysin (alpha toxin) production by lysing human blood while 30 (63.8%) phenotypically expressed biofilm production. The difference in the gender distribution of the volunteers with haemolysin producing S. aureus isolates was not statistically significant (Odds ratio=1.799; χ2=2.857; p=0.091). However, biofilm producing S. aureus isolates were significantly higher in the males than the females (Odds ratio=2.394; χ2=5.474; p=0.019) [Table/Fig-4].
The gender distribution of volunteers with haemolysin and biofilm producing nasal S. aureus.
| Virulent factor | Detection | No. of sample | Females | Males | Chi-square | p-value |
|---|
| Haemolysin | Yes | 37 | 21 | 16 | 1.799 | 0.091 |
| No | 363 | 255 | 108 |
| Biofilm | Yes | 30 | 15 | 15 | 5.474 | 0.019* |
| No | 370 | 261 | 109 |
*Statistically significant (p<0.05)
Antimicrobial Resistant Patterns of S. aureus and MRSA Isolates
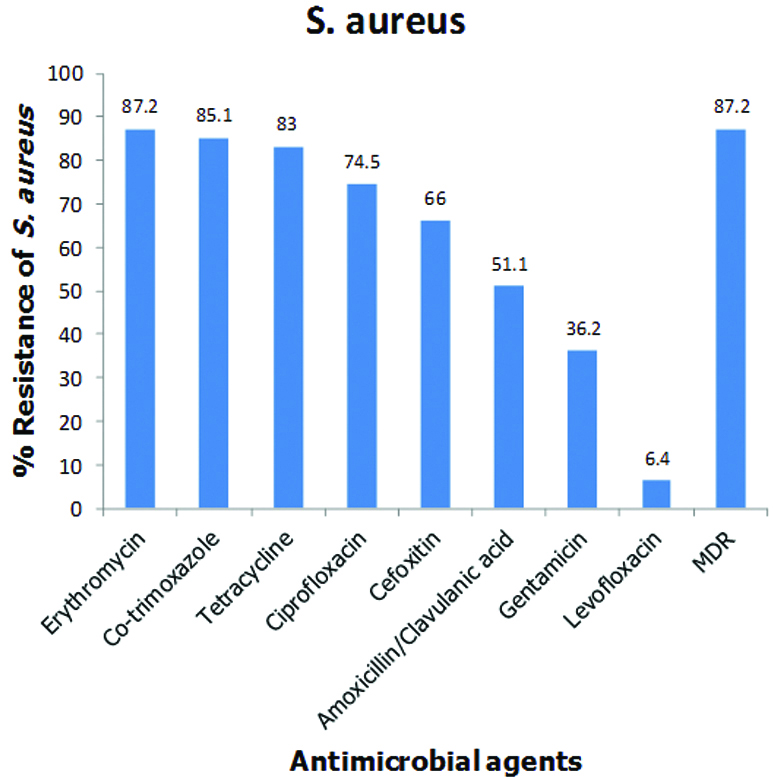
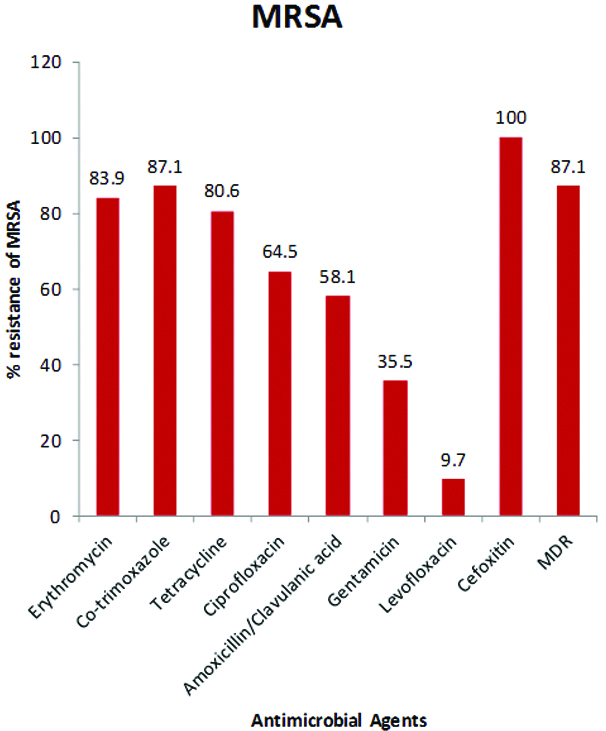
The antimicrobial susceptibility testing of the S. aureus isolates revealed that 31 (66%) of them phenotypically expressed methicillin resistance being moderately resistant to cefoxitin (66%), an agent approved by CLSI for the phenotypic screening for methicillin resistant S. aureus (MRSA). Hence, 31 (66%; 31/47) of the isolates were MRSA while the overall nasal carriage rate of MRSA among all the healthy volunteers was 31 (7.8%; 31/400). No mecA gene was detected in all the 47 S. aureus isolates screened by PCR analysis. The isolates also exhibited moderate resistance to amoxicillin/clavulanic acid (51.1%) and gentamycin (36.2%) but high resistance to erythromycin (87.2%), co-trimoxazole (85.1%), tetracycline (83%) and ciprofloxacin (74.5%). However, very low resistance was exhibited to levofloxacin (6.4%). The biofilm producers in this study were found to be resistant to at least two antimicrobial agents. The antimicrobial resistance profiles of all the 47 S. aureus and 31 MRSA isolates are shown in [Table/Fig-5,6] respectively.
Antimicrobial resistant profile of nasal S. aureus isolates.
Antimicrobial resistant profile of nasal MRSA isolates.
Prevalence and Antibiogram of MDR
The prevalence of multidrug resistance among the isolates was 41 (87.2%) and 27 (65.9%) of them were MRSA. A total of 16 types of antibiogram of the antimicrobial agents were observed among the multidrug resistant S. aureus isolates and the most predominant antibiotic combinations among the multi-drug resistant isolates were “E, SXT, TE” (35, 85.4%), CIP, SXT, TE” (31, 75.6%), “CIP, E, SXT, TE” (28, 68.3%) and “CN, CIP, E, SXT, TE” (14, 34.1%) as shown in [Table/Fig-7]. Thirty-three (33/41; 80.5%) and 26 (26/41; 63.4%) of the MDR isolates were haemolysin and biofilm producers respectively.
The Antibiogram of multi-drug resistant Staphylococcus aureus.
| Combination of antimicrobial agents to which the isolates exhibited multidrug resistant | Number of resistant isolates |
|---|
| AMC, E, TE | 1 |
| FOX, CIP, SXT | 1 |
| AMC, CIP, SXT, TE | 1 |
| FOX, E, SXT, TE | 2 |
| FOX, CIP, SXT, TE | 2 |
| CIP, E, SXT, TE | 4 |
| AMC, FOX, E, SXT, TE | 3 |
| FOX, CN, CIP, E, SXT | 1 |
| FOX, CIP, E, SXT, TE | 2 |
| CN, CIP, E, SXT, TE | 4 |
| AMC, CIP, E, SXT, TE | 2 |
| AMC, FOX, CIP, E, SXT, TE | 6 |
| AMC, CN, FOX, E, SXT, TE | 2 |
| AMC, CN, CIP, E, SXT, TE | 2 |
| FOX, CN, CIP, E, SXT, TE | 1 |
| AMC, FOX, CN, CIP, E, SXT, TE | 7 |
| Total | 41 (87.2%) |
AMC: Amoxicillin/Clavulanic acid; CIP: Ciprofloxacin; CN: Gentamycin; E: Erythromycin, FOX: Cefoxitin; SXT: Co-trimoxazole; TE: Tetracycline
Discussion
Nasal carriage of S. aureus has been reported to be a significant risk factor for the development of nosocomial and community-acquired infection in a variety of populations [2,21,22]. The rapid emergence of multidrug resistant strains among this organism especially those that possess mecA gene which encodes for methicillin resistance has made the treatment of S. aureus infections difficult because of limited treatment options [23,24]. Hence, regular surveillance of the pathogenic and resistance potentials of this organism in this environment where there is little or no control on the use of antibiotics is needed for adoption of strategies for healthy society.
The asymptomatic nasal carriage rate of S. aureus has been widely reported in relevant studies to be around 20-30% [2-4]. The nasal carriage rate in this study (11.8%) was however smaller than the general range but it is similar to the recent reports of Omuse G et al., in Kenya (18.3%), Ben-Slama K et al., in Tunisia (13%), and Tigabu A et al., in Ethiopia (13.3%) [10,25,26]. In contrast to our results are the findings from Gabon (29%) [27], Netherlands (25.2%) [28] and Norway (27.6%) [29]. The nasal carriage rate of S. aureus in healthy population has been observed to depend on the geographical location, age of the subjects and the screening techniques [21,30]. Thus, differences across the various study centres might be due to these observations. Also, the observed significantly lower prevalence of nasal carriage of S. aureus among the female students (p=0.013) might be due to their higher consciousness of hygiene practices than the males which help to reduce the transfer of this organism from other parts of the body to the nostrils.
The investigation into the capacity of the studied S. aureus isolates to phenotypically expression haemolysin production as a characteristic virulence trait, revealed 37 (78.7%) isolates to have secreted alpha-haemolytic toxin having lysed the red blood cells. Staphylococcus aureus α-haemolysin is known to possess Hla gene on its chromosome which encodes for the formation of a complete beta-barrel pore-forming cytotoxins responsible for the development of pores in the cellular membrane that eventually cause cell death [5,31]. An increased Hla gene expression in S. aureus alpha-toxin can cause a significant disease burden in healthy humans intoxicating multiple human cell types ranging from epithelial cells, endothelial cells, and an array of other haematopoietic-lineage cells [31]. Hence, the high prevalence of these strains among the asymptomatic carriers of nasal S. aureus in this study can lead to increase morbidity and mortality as well as healthcare costs due to opportunistic infections and its transmission to the entire society [32]. The difference in the prevalence of the haemolysin producing S. aureus among the genders was however not significant. This therefore suggests that no activity of the males predisposes them more to haemolysin production than the females.
A prevalence of 30 (63.8%) isolates was found to phenotypically expressed biofilm production in this study. This result is similar with the findings in Nepal (61%) by Devapriya F et al., and in Pakistan (54.8%) by Wroblewska J et al., from healthy volunteers using the same conventional screening method [33,34]. Similar screening for biofilm formation among nasal S. aureus in healthy populations is very scarce in Nigeria and other Africa countries. This observed high prevalence of biofilm producing nasal S. aureus in this study is a major public health problem since biofilm has been known to exhibit protective strategies against the host defense mechanism and restrict the penetration of some antimicrobial agents causing recurrent infections that are irresponsive to treatments [6,8,21,33]. The prevalence of biofilm producing S. aureus isolates was however significantly higher in the males than the females (p=0.019) and this might be due the relatively lower practices of personal hygiene among the male students than the females. This study also revealed a high proportion of 22 (59.5%) biofilm producers among the isolates that expressed the secretion of alpha-haemolytic toxin and this supports the finding of Anderson MJ et al., which reported that alpha-toxin promote the formation of S. aureus mucosal biofilm [35]. It was therefore suggested that strategies to neutralise alpha-toxin in S. aureus could be a potent therapeutic target for some of its infections that are difficult to treat [35].
The antimicrobial susceptibility screening showed the isolates to be highly resistant to erythromycin, co-trimoxazole, tetracycline and amoxicillin/clavulanic acid in this study. Many recent studies have reported similar findings among nasal S.aureus isolates from healthy adults in Nigeria and other developing countries [23,36-38]. The heavy and uncontrollable usage of these agents in both human and agricultural activities in the community is likely to be responsible for the organism’s high resistance to them [38]. A comparison of this present result with the findings of Onanuga A et al., in a similar study in the same environment five years ago, in which nasal S. aureus from healthy volunteers exhibited lower resistance to amoxicillin/clavulanic acid (5%), cotrimoxazole (32.5%) and erythromycin (35%), shows the rapid increasing rate (100-350%) at which this organism develops resistance within the same environment [39]. This therefore suggests the increasing antibiotic pressure in the environment which favours the observed increased antibiotic resistance over the short periods of time [40].
The prevalence of MRSA, a pathogenic strain of S. aureus that is known to cause difficult to treat infections in both hospital and community settings worldwide, has been widely reported in nasal cavity of healthy volunteers. Our study detected 31 (66%) of the isolates to be cefoxitin resistant, a measure of the phenotypic methicillin resistant (MRSA). Thus, the overall nasal carriage rate of MRSA among all the healthy volunteers was 31 (7.8%). Higher MRSA prevalence among isolates from healthy volunteers has been reported by Owolabi JB et al., in Ota, Nigeria (100%) and El Aila NA et al., in Gaza-Palestine, Israel (82.3%) [38,41]. However, lower prevalence has also been reported among isolates from similar subjects by Shinde RV et al., in India (34.6%), Akerele JO et al., in Benin-City, Nigeria (22%), Ugwu MC et al., in Agbor, Nigeria (25.7%), and Shibabaw A et al., in Northeast Ethiopia (44.1%) [24,42-44]. The observed differences might be due to the different populations and their exposure to antibiotics and other risk factors.
The cefoxitin-resistant S. aureus isolates in this study did not possess mecA gene that code for MRSA. This observed absence of mecA gene in phenotypic MRSA has been reported by El Aila NA et al., in Gaza-Palestine, Israel, Olayinka BO et al., in Zaria, Nigeria, and Elhassan MM et al., in Sudan [41,45,46]. There have been reports of a recently recognised form of MRSA encoding a divergent mec gene called mecC MRSA in European countries, which colonise and cause disease in humans and a wide range of other host species [47]. However, the screening for mecC gene among the isolates was not within the scope of this study. Hence, the observed resistance in these isolates might be due to the possession of mecC which was not investigated in this study. Furthermore, the types of isolates that do not have an altered PBP2a, encoded by the mecA or mecC gene have also been described as Borderline oxacillin-resistant Staphylococcus aureus (BORSA) and their resistance has been typically associated with hyperproduction of beta-lactamases or point mutations in PBP genes [48,49]. The clinical presentation and epidemiology of these BORSA infections are similar to those of MRSA and are found in both hospital and community settings [45,48]. Hence, the increasing emergence of mecC MRSA and BORSA in the phenotypic MRSA that lack mecA gene demands their frequent surveillance in our environments.
The S. aureus and MRSA isolates in this study exhibited very high susceptibility to levofloxacin and gentamycin but high resistance was observed to ciprofloxacin. This findings have been reported in many recent studies [38,42,50]. The higher ciprofloxacin resistance (74.5%) among these isolates in this environment where 10% were resistant among similar subjects five years ago is alarming, since ciprofloxacin in the last decade was an alternative for the treatment MRSA infections. This observed increased resistance to ciprofloxacin relative to levofloxacin might be due to its over-prescription for virtually all bacterial infections without recourse to culture and susceptibility testing of the causative agent which leads to the proliferation of its different brands with cheaper prices in the market compared to levofloxacin which is not often prescribed. Thus, this might lead to selective pressure on the organism due to the vast inappropriate and uncontrollable usage in the community. Gentamycin still maintains its high activity on the organism (though with low increase in resistance) possibly because it is an injectable which is dislike by many individuals.
The prevalence of MDR among the isolates was 41 (87.2%) and 27 (65.9%) of them were MRSA. Also, 33 (33/41; 80.5%) and 26 (26/41; 63.4%) of the MDR isolates were haemolysin and biofilm producers respectively suggesting higher virulence capability of the MDR strains. Prominent among the MDR isolates are the E, SXT, TE (85.4%) and CIP, SXT, TE (75.6%) antibiogram which suggest that the isolates have been majorly exposed to erythromycin, co-trimoxazole, tetracycline and ciprofloxacin in the community which therefore indicates that these agents have been grossly misused by the study subjects in this environment thus increasingly limiting the therapeutic options for infections caused by these S. aureus strains. This situation calls for urgent strict antibiotic control and stewardship in the community.
Limitation
This study has some limitations which are: the phenotypic detection of MRSA isolates were carried out only with cefoxitin disc and the non investigation of mecC gene among the phenotypic MRSA isolates that lack mecA gene. Thus, the use of both cefoxitin and oxacillin discs in the detection of phenotypic MRSA and the screening for mecC gene among S. aureus isolates are recommended for future studies since there is an increasing emergence of the mecC form of MRSA in the world.
Conclusion
This study revealed a high prevalence of MDR isolates with high proportions of haemolysin and biofilm producers among the healthy University students. This therefore calls for prompt implementation of proper personal hygiene strategies through regular daily hand washing in order to prevent the spread of these virulent MDR strains and strict antibiotic stewardship for the control of MDR in the community.