The purpose of this study was to detect hVISA and VISA isolates among MRSA and MSSA strains obtained from various clinical samples as vancomycin remains the drug of choice to treat MRSA infections and also to study the phenotypic and genotypic characteristics of these isolates.
Materials and Methods
Study type and collection of S.aureus strains: The present descriptive study was carried out in Department of Microbiology, Chettinad Hospital and Research Institute, Kelambakkam, Chennai, Tamil Nadu, India. A total of 468 non-duplicate S.aureus strains, obtained from various clinical specimens including blood, exudates, urine and respiratory samples during a period of two years from September 2014 to November 2016 were included in the study. This study was approved by Institutional Human Ethics Committee, CARE (IHEC/02/2014/ D.No.322). These strains were stocked in 20% glycerol broth and stored at -20°C until used for further analysis.
Identification of S.aureus strains:S.aureus strains from various clinical specimens were identified by standard microbiological procedures which included routine Gram stain demonstrating Gram positive cocci in clusters and positive catalase test, tube coagulase test and mannitol fermentations tests [12,13].
Detection of hVISA/VISA by screening method: All the 468 S.aureus strains including both MRSA and MSSA were screened for hVISA/VISA by using Brain Heart Infusion Agar (BHIA) containing 6 μg vancomycin [14, 15]. A 10 μL of 0.5 McFarland suspensions of the strains were spot inoculated in BHIA with 6 μg vancomycin. The plates were incubated for 24 hours at 35°C±2°C and inspected for the growth of any single colony of S.aureus strain. Screening test was done in triplicates.
Confirmatory test population analysis profile area under the curve (PAP-AUC):S.aureus strains that were screened positive for hVISA/VISA were further subjected to PAP-AUC method which remains the gold standard method to confirm hVISA/VISA isolates and Mu3 (ATCC 700698), Mu 50 (ATCC 700699) were used as control strains [16]. Briefly, overnight broth cultures of S.aureus were diluted to obtain 10-3, 10-6, 10-8 and this was manually spiral plated on BHIA containing 0.25 μg/mL, 0.5 μg/mL, 1.0 μg/mL, 1.5 μg/mL, 2 μg/ml, 4 μg/mL, 6 μg/mL, 8 μg/mL and 16 μg/mL of vancomycin.
The colonies were counted in each plate at the end of 48 hour of incubation at 37°C. The viable count of S.aureus strains was plotted against vancomycin concentration on a logarithmic paper. This was used to calculate the AUC. The ratio of AUC of the test strain to that of the Mu3 (ATCC 700698) was obtained to distinguish between VSSA, hVISA and VISA. A ratio of ≥0.9-1.2 was used to diagnose hVISA and a ratio of >1.3 for VISA [16].
Triton X induced autolytic assay: Triton X was used to stimulate autolysis as hVISA/VISA strains have thickened cell wall due to defective synthesis of murein hydrolase responsible for the regulation of cell wall turnover. Briefly, S.aureus isolates were grown in BHI broth overnight for 14-18 hours (mid log phase cells). The bacterial cells were pelleted and washed twice in ice-cold distilled water and resuspended in 0.05% Triton X, incubated at 37°C and OD 600 values were obtained every 30 minutes. Percentage of lysis was calculated by dividing OD by initial OD x 100 [17].
Colony spreading assay: This assay was carried out in Trypticase soy agar containing 0.24% agar. The plates were dried in safety cabinet for 20 minutes and about 2 μL of 0.5 McFarland bacterial suspensions was spot inoculated and dried in the safety cabinet for another 15-20 minutes. Then the plates were incubated for 10 hours at 37°C and after incubation, the colonies were inspected for spreading over the agar surface [18].
TEM analysis: To confirm cell wall thickness for hVISA/VISA strains Transmission Electron Microscopy (TEM) analysis was done.
Detection of virulence factors for hVISA/VISA/MRSA strains:
Slime formation: Slime formation was detected in MRSA/hVISA/VISA strains using Congo Red Agar (CRA) containing BHI broth, 5% sucrose, agar and 0.08% congo red. Strains that grew as dry black colonies with crystalline consistency were considered as slime producers and those that produced pink colonies were considered as negative for slime formation [11,15] [Table/Fig-1].
Slime production demonstrated in 5% sucrose containing congo red agar. Slime producing strains appear as dry black colonies.

Biofilm assay: Biofilm assay was carried out for MRSA/hVISA/VISA isolates according to Kwasny SM et al., and Stepanovic S et al., [19, 20]. Briefly, 200 μL of the bacterial suspension incubated overnight was transferred to 96 wells flat bottomed microtitre plate containing 200 μL of Brain Heart Infusion Broth (BHIB) containing 1% glucose. After 24 hours of incubation, the plates were removed and quantified for overall growth at OD550. Then the plates were washed three times in distilled water using plate washer.
The biofilm were fixed by incubating the washed plate at 60°C for 1 hour. The biofilm were stained using 0.06% crystal violet for 5 minutes and washed thrice with distilled water using plate reader. Quantification of biofilm was done by eluting the bound crystal violet with 30% acetic acid and OD 550 was measured. The cut-off OD was calculated as three standard deviations above the mean OD of the negative control. Strains were non-adherent if the OD≤ODc [15,20].
Haemagglutination: Haemagglutination test was carried out in a 96 well round bottom microtitre plate. The bacterial colony was suspended in Phosphate Buffer Saline (PBS) and equal volume of 1% O group RBC’s was added and haemagglutination was observed after 2 hours of incubation [11,15].
Detection of DNase: This test was carried out by using DNase agar (Himedia Laboratories Ltd.,). The bacterial strains were considered positive for DNase if clearing around the spot inoculated colony was observed after pouring 1N HCl [15,21].
Detection of beta haemolysin: The strains were spot inoculated onto 5% sheep blood agar and incubated at 37°C overnight. After overnight incubation, the plates were kept at 4°C to observe hot-cold type of hemolysis produced by beta haemolysin [15,22].
Quantitative assay for DNase and haemolysins: The strains were grown in BHI broth for 24 hours. Bacterial cells were pelleted at 10,000 rpm and supernatant containing the haemolysins and DNase were obtained. The supernatant (1 mL) was subjected to doubling dilution and 10 μL suspension from each dilution was added into appropriate wells cut in 5% sheep blood agar and DNase agar for estimating hemolysin and DNase production by the strains. After overnight incubation the zone of haemolysis and zone of clearing for DNase production were measured in mm. Strains showing consistently larger zones sizes till 1:16 dilution were considered as abundant producers of haemolysin and DNase.
Detection of antibiotic resistance pattern of hVISA isolates: Kirby Bauer disc diffusion method was done to detect the susceptibility pattern of hVISA/VISA strains to various antibiotics. Minimum inhibitory concentration for mupirocin was also detected for hVISA strains by agar dilution method according to CLSI guidelines. Fusidic acid susceptibility test was carried out for High Level Mupirocin Resistant (HLMR) isolates.
Detection of Agr specificity for hVISA and VISA strains
DNA extraction: DNA from hVISA/VISA strains was extracted by microwave method. The bacterial suspension were microwaved intermittently for two minutes and centrifuged at 10,000 rpm. Supernatant was used as DNA template.
Agr specific PCR: The agr specificity for hVISA/VISA strains was detected by amplifying the hyper variable region of the agr locus using oligonucleotide primers (one forward pan agr and four reverse primers) which is specific for the four agr types. This includes, forward pan-agr primer-5’-ATGCACATGGTGCACATGC-3’ Reverse-agr I 5’-GTCACAAGTACTATAAGCTGCGAT-3’, Reverse- agr II-5’GTATTACTAATTGAAAAGTGCCATAGC-3’, Reverse-agr III-5’-CTGTTGAAAAAGTCAACTAAAAGCTC-3’, Reverse-agr IV-5’-CGATAATGCCG TAATAC CCG-3’.
Since the product size of I (440 bp) and III (406 bp) and II (572 bp) and IV (588 bp) were similar, two duplex PCR reactions were carried out. The PCR conditions include: 1 cycle of initial denaturation at 95°C for 5 minutes, followed by 30 cycles of denaturation (94°C for 1 minutes), annealing (55°C for 1 minute), elongation (72°C for 1 minute) and final elongation (72°C for 5 minutes). The amplicons were analysed by using 1% agarose electrophoresis [23, 24].
Statistical Analysis
The results were analysed using IBM SPSS (version 21.0) software. The prevalence of hVISA among S.aureus strains and the prevalence of hVISA among MSSA and MRSA strains were analysed by proportion test. The characters of hVISA strains were analysed using chi-square test. A p-value of <0.05 was considered as statistically significant.
Results
Prevalence of hVISA strains by screening and confirmatory method: Of the 468 S.aureus strains, 77 (16.4%) strains were screened positive for hVISA by 6μg vancomycin screen agar. However, out of these 77 hVISA/VISA strains only 33 (7%) of the strains were confirmed as hVISA by PAP-AUC method which remains as one of the gold standard technique for confirming hVISA strains. All the strains had PAP-AUC ratio between >0.9-1.2. [Table/Fig-2] Among hVISA, 11 strains were MRSA (33%).
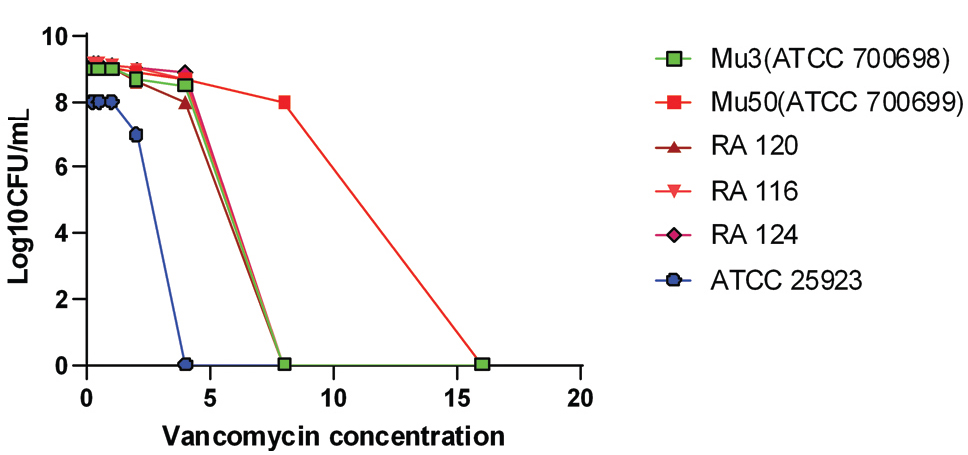
Shows population analysis of control strains ATCC 25923 (VSSA), ATCC700698 (Mu3-hVISA), ATCC700699 (Mu50-VISA) and representative test strains (RA116, RA120 and RA124). Total number of colonies grown on Brain heart infusion agar containing varying concentration of vancomycin that was counted after 48 h of incubation. Positive control strains Mu50, Mu3 and test strains RA 116, RA120 and RA124 shows reduced susceptibility to vancomycin (>4 μg/mL) and negative control strain ATCC 25923 shows increased susceptibility to vancomycin (<4 μg/mL).
Of the total 468 S.aureus strains analysed in the present study, 11 (2.3%) of the hVISA isolates were MRSA and 22 (4.7%) of the hVISA isolates were MSSA. Out of the 114 MRSA strains, 11 (9.65%) strains were hVISA and out of 354 MSSA 22 (6.2%) were hVISA strains. Of the 33 hVISA strains detected, 11 (33.3%) of them were MRSA and the remaining 22 (66.6%) of the hVISA strains were MSSA isolates.
The prevalence of hVISA among S.aureus isolates were found to be significant (p<0.001). Similarly the prevalence of hVISA among MRSA isolates were found to be significant (p<0.001). The prevalence of hVISA among MSSA isolates were found to be significantly lower when compared to MRSA isolates.
Among the 33 of the hVISA isolated, 5 (15%) were from blood, 1 (3%) were from urine and 27 (81%) of the isolates were from exudates samples. Of the 23 blood, 24 urine, 13 respiratory and 408 exudate S.aureus strains, 21.7%, 4.2%, 0% and 6.6% of them were hVISA strains respectively. hVISA strains were prevalent among blood isolates (invasive).
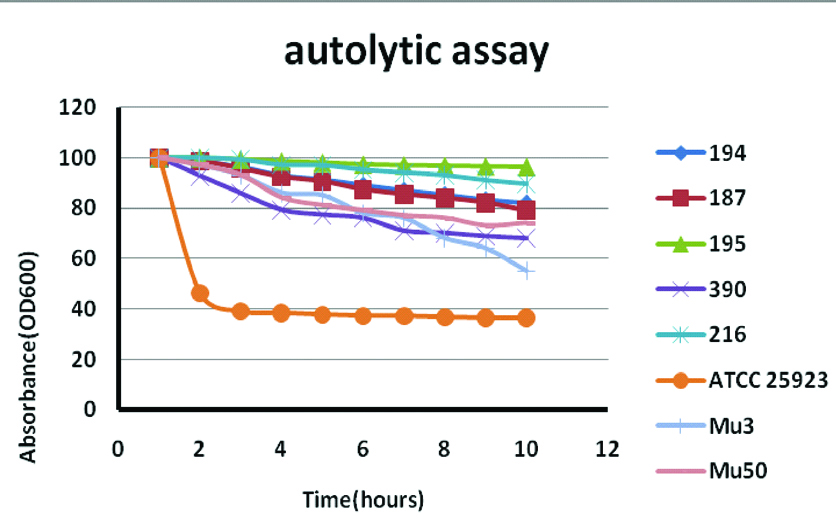
All the 33 hVISA strains (100%) showed reduced autolytic activity with triton-X 100 induced lysis and reduced colony spreading property was appreciated in 13(39%) of these isolates. Autolytic assay absorbance represented as % absorbance is depicted in [Table/Fig-3].
Autolytic assay with Triton X-100 induced lysis for control strains (Mu3 and Mu50) and representative test strains (RA194, RA187, RA195, RA390, RA216). Triton X 100 induced autolytic assay Absorbance represented as % of absorbance (OD 600nm). ATCC 25923 (VSSA) showed increased autolysis within 2 hours whereas positive control strains and test strains showed reduced autolysis up to 10 hours of incubation.
Transmission Electron Microscopy Analysis of hVISA Strain
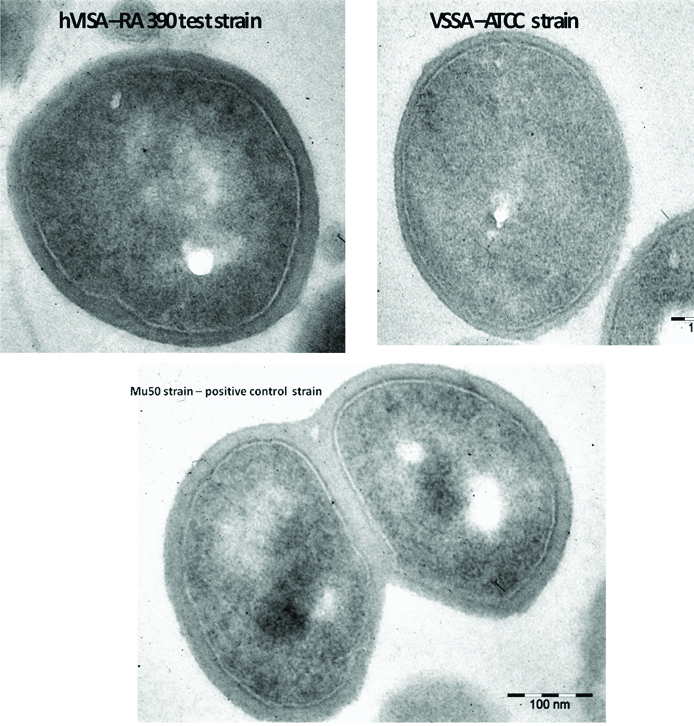
Transmission Electron Micrograph analysis of represented isolates shows significant thickness of cell wall [Table/Fig-4]. Test strain RA 390 (hVISA) and Mu 50 (VISA) showed a significant increase in cell wall thickness when compared to ATCC 25923 (VSSA).
TEM analysis-hVISA- RA 390 (test) strain showing thick cell wall (Magnification- 25,000X) Cell wall thickness-31.76 nm, VISA Mu 50 strain showing Cell wall thickness-47.39 nm and VSSA -ATCC 25923 strain showing cell wall thickness-13.51 nm.
Virulence Properties of hVISA Strains
Among the hVISA strains, 14 strains (42.4%) were slime producers, 15 strains (45.4%) were biofilm producers, 6 (18%) were negative for DNase, 2 (6%) were weakly positive for coagulase, 25 (76%) were positive for haemolysin, 18 (55%) agglutinated 1% O group RBC’s and 13 (39%) did not have colony spreading. Quantitative assay revealed only 42% (14) and 39% (13) isolates as strong producers of DNase and haemolysin respectively.
Few hVISA isolates had significantly greater coagulation time and less DNAase production when compared to VSSA isolates (p<0.001). Slime formation and biofilm production were found to be significantly higher in hVISA isolates (p=0.03 and p <0.001 respectively). However haemagglutination and haemolysin production did not show any significance among hVISA isolates and VSSA isolates (p=0.283 and 0.192 respectively). There was a greater association between the thicker cell wall as demonstrated by Triton X 100 induced lysis and TEM analysis with reduced coagulase and DNAse production (p<0.001).
Antibiotic resistance pattern of hVISA isolates: hVISA isolates showed significantly less resistance to various group of antibiotics other than vancomycin, like netilmycin (0%) clindamycin (3%), tetracycline (3%), gentamycin (12%), cloxacillin (33%), erythromycin (36%), ciprofloxacin (36%) and cotrimoxazole (39%). High level mupirocin resistance was observed in 6.06 % of the hVISA strains. All the HLMR isolates were susceptible to fusidic acid.
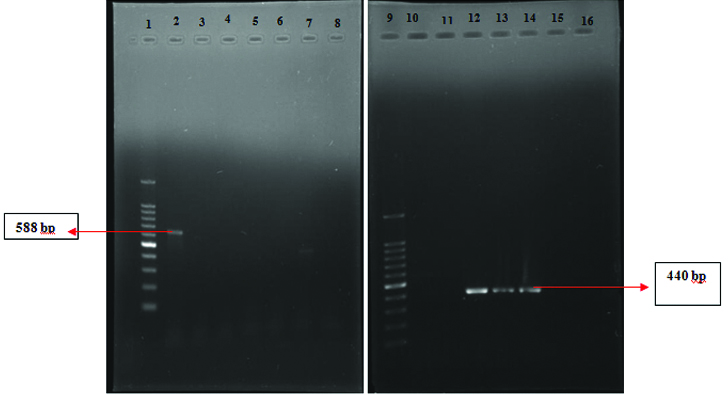
Agr duplex PCR: Out of the 33 hVISA strains 14 (42.4%) belonged to agr I (440bp), 8 (24.2%) belonged to agr II (572bp), 10 (30%) belonged to agr III (406 bp) and 1(3%) belonged to agr IV (588bp). [Table/Fig-5,6].
Agr Duplex PCR-agr type II and III (L1-100bp Ladder, L2-agr II, L3-Nil, L4-agr III , L5 agr II, L6 agr II, L7 & 8-Nil).
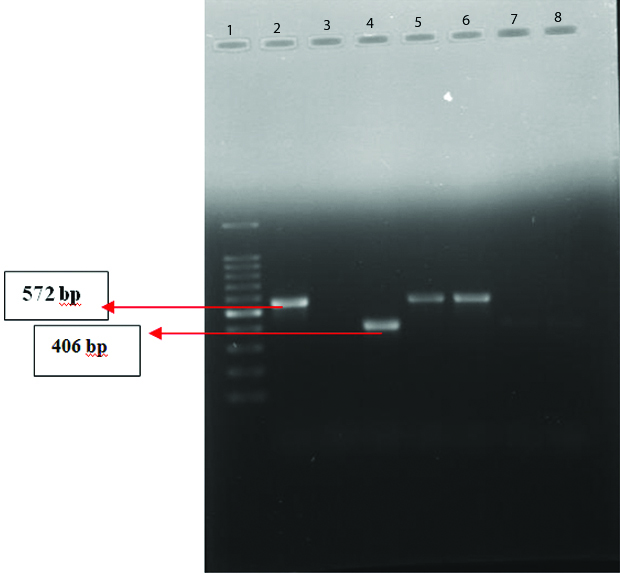
Agr Duplex PCR agr type I and IV (L1-100 bp ladder, L2-agr IV, L12,13,14-agr I, L3 to L8, L10 & 11, L15 & 16-Nil).
Discussion
In this study, hVISA prevalence rate was 7% (33) and this is consistent with a study by Chaudhari CN et al., from India that shows hVISA/VISA prevalence rate of 6.9% and Asian Network for Surveillance of Resistant Pathogens (ANSORP) gives a prevalence rate of 6.3% [25,26]. The hVISA/VISA strains among MRSA isolates were found to be 33%, whereas 66% of the hVISA/VISA isolates were MSSA isolates. This highlights the fact that reduction in vancomycin susceptibility has become common among MSSA [27]. Therefore, treatment of MSSA isolates with vancomycin can lead to treatment failures [3,4].
Strains of S.aureus which form biofilm display antibiotic resistance and colonize most of the indwelling medical devices [28,29]. In the present study, all the hVISA/VISA isolates showed reduced autolytic activity with Triton-X induced lysis, increased cell wall thickness by TEM and 13 (39%) of these isolates did not show colony spreading. 6 (18%) were negative for DNase, 2 (6%) were weakly positive for coagulase, 15 (45%) were biofilm producers and only 25 (76%) were positive for haemolysin production. This explains that these isolates had thickened cell wall and therefore non-susceptible to triton-X induced lysis [30]. Decreased autolytic activity and colony spreading among hVISA isolates can be due to reduced agr function.
Agr was found to up regulate extracellular virulence factors like DNase, coagulase, etc., and also responsible for colony spreading on soft agar surface. Thus reduced colony spreading, reduction in DNase, coagulase suggests agr dysfunction in these isolates [10,31]. Late coagulase production can be due to the thickened cell wall and this can lead to misidentification of the S.aureus isolates.
In the present study, out of the 33 hVISA/VISA strains 14 (42.4%) belonged to agr I, 8 (24.2%) belonged to agr II, 10 (30%) belonged to agr III and 1 (3%) belonged to agr IV. A study by Singh A et al., shows that 82.8% were agr type I, 3/29 10.3% were type III and 6.9% were non-typeable [32]. However, in the present study all the strains were typeable by agr duplex PCR. Among Methicillin Resistant-hVISA agr type II was found to be the predominant type and this was concordant with many previous studies [33, 34].
The hVISA/VISA isolates showed better susceptibility pattern to other group of antibiotics like aminoglycosides, macrolides, quinolones, cotrimoxazole and tetracycline when compared with MRSA isolates. Hence judicious use of antibiotics is recommended in healthcare settings in order to avoid the emergence of multidrug resistant strains. Therefore, overuse of vancomycin can create selective pressure and may lead to the emergence of strains with reduced vancomycin susceptibility.
Limitation and Future Recommendation
In this study few characteristics of hVISA strains like the agr dysfunction, agr polymorphism and some phenotypic properties were analysed. However, mutations of several regulatory genes like vraSR, walkR, graR and rpoB can be associated in regulation of vancomycin intermediate resistance. Therefore, analysis of these genes by sequencing can further help to discern the complex mechanisms that underlie vancomycin intermediate resistance. Further a prospective study can be done to analyse the predictive elements for the increase in vancomycin MIC like recent exposure to vancomycin or MRSA bacteraemia because vancomycin overuse can create selective pressure leading to emergence of hVISA/VISA clones.
Conclusion
S.aureus infections have become difficult to treat and life-threatening due to increase in the number of strains with reduced susceptibility to vancomycin. In the current study, all the isolates detected as hVISA/VISA had diverse phenotypic characteristics and gave vancomycin MIC’s <1.5 μg/mL by routine CLSI method. These strains were detected in both MRSA and MSSA isolates and the shift is due to the overuse of vancomycin in healthcare settings to treat both MRSA and MSSA infections. Therefore, this stresses the importance of detecting these isolates in healthcare settings and prudent use of vancomycin can prevent treatment failures in future.